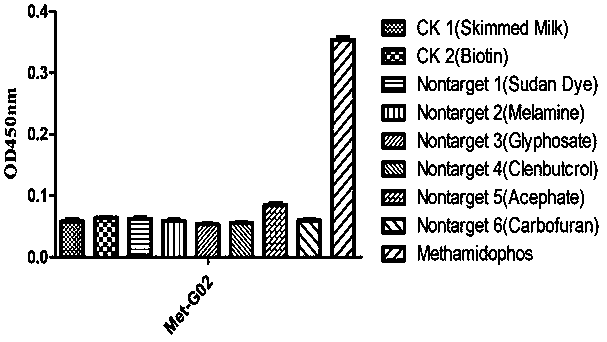
Aptamer Met-G02 specifically combined with methamidophos and application thereof
A nucleic acid aptamer, methamidophos technology, which is applied in the field of biomedicine and can solve problems such as inability to be widely used
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1: Screening of methamidophos nucleic acid aptamer
[0039] 1. Coupling of Ligand (Methamidophos) and Matrix (Sepharose 6B)
[0040] 1. Suspend 1 g of freeze-dried powder Sepharose 6B in 3 mL of distilled water in a sintered glass filter (porosity G3);
[0041] 2. Immediately wash the matrix with 200 mL of coupling buffer solution (0.05 M, pH9.6 carbonate buffer) in a sintered glass filter for 1 h;
[0042] 3. Dissolve 6 g of ligand with 6 mL of coupling buffer solution to make the final concentration 1 mg / mL;
[0043] 4. Mix the substrate in step 2 with the buffer solution in step 3 with a concentration of 1 mg / mL in which the ligand has been dissolved in a ratio of 1:2 by volume, and treat it for 16 h under the condition of a water bath at 35°C-40°C, and 37 h ℃ shaker overnight;
[0044] 5. At 40°C-50°C, block excess groups with 1M aminoethanol for at least 4 hours or overnight;
[0045] 6. Wash the excess ligand with coupling buffer, centrifuge at 4000...
Embodiment 2
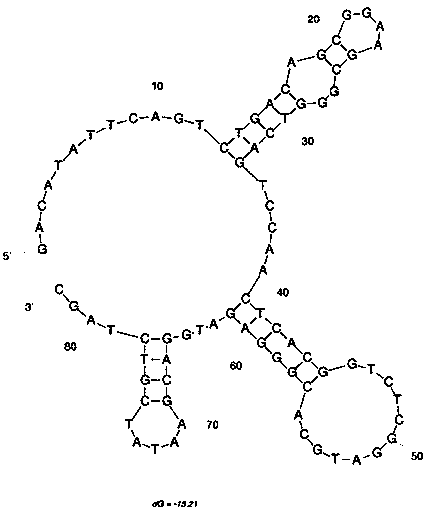
[0090] Example 2: Cloning, isolation and sequencing of nucleic acid aptamers and prediction of secondary structure of single-stranded DNA
[0091] 1. Preparation of Escherichia coli DH5α Competent Cells
[0092] 1. Pick a single DH5α colony, inoculate it in 3 mL of LB medium without ampicillin, and incubate overnight at 37 °C. The next day, take the above bacterial solution and inoculate it in 50 mL of liquid LB medium at a ratio of 1:100, 37 Shake at ℃ for 2 min; when the OD600 value reaches 0.35, harvest the bacterial culture;
[0093] 2. Transfer the bacterial culture to a 50 mL pre-cooled sterile polypropylene tube and place it on ice for 10 min to cool the culture;
[0094] 3. Centrifuge at 4000 rpm for 10 minutes at 4°C, discard the culture medium, and invert the tube for 1 minute to drain the remaining culture medium;
[0095] 4. Add 150 μL ice-cooled 0.1 mM CaCl each 2 Solution, combine two tubes, ice bath for 10 min;
[0096] 5. Centrifuge at 4000 rpm for 10 min a...
Embodiment 3
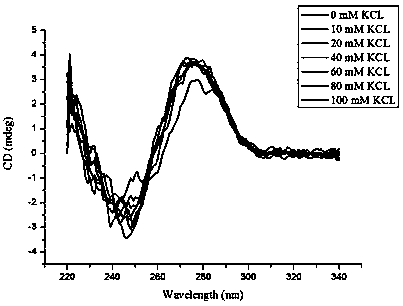
[0110] Embodiment 3: circular dichroism to K + Research on the relationship between concentration and nucleic acid aptamer Met-G02
[0111] 1. Dilute the aptamer with sterilized water to 20 μM, denature at 94 °C for 0.5 min, and cool to 25 °C at a rate of 0.5 °C / min;
[0112] 2. Dilute the nucleic acid aptamer Met-G02 to 2.5 μM with KCl solutions of different concentrations (0, 10, 20, 40, 60, 80, 100 mM);
[0113] 3. Detect with a circular dichroism spectrometer at 25 °C and a wavelength of 220–340 nm (see the results in figure 2 ).
[0114] The results showed that the chromatograms had negative peaks between 240-250 nm and positive peaks between 275-285 nm. Because the peak at 240-250 nm is the characteristic peak of G-quadruplex and the peak at 275-285 nm is the characteristic peak of the stem-loop, the nucleic acid aptamer Met-G02 can exist stably in KCl solution without affecting its secondary structure .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com