Method for constructing hESC indicating cell line for specifically tracing endothelial cell differentiation
A technology for indicating cell lines and endothelial cells, which can be used in biochemical equipment and methods, embryonic cells, animal cells, etc., and can solve problems such as lack of regulatory elements, unstable expression, and position effects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1 Experimental process
[0045] The primers and probes to be used are listed in Table 1:
[0046] Table 1
[0047]
[0048]
[0049] (1) CD144N-GFP vector construction
[0050] ① Amplify the 5′ and 3′ homology arms, using H9 gDNA as a template:
[0051] The PCR system is as follows:
[0052]
[0053] The thermal cycling conditions are:
[0054]
[0055]
[0056] The primer used for the 5' homology arm is CD144N5F2+CD144N5Rovr, and the product is 831bp.
[0057] The primers used for the 3' homology arm are CD144N3F+CD144N3R1, and the product is 568bp.
[0058] ② Amplify orf+pa of GFP from pEGFP-N1, and add restriction sites of Xba 1 and Nhe 1 in the upstream and downstream respectively. The PCR system is as follows:
[0059]
[0060] The thermal cycling conditions are:
[0061]
[0062] Using CD144N-GFP-ovF+CD144N-GFP-R as primers, the product size is 961bp
[0063] ③ Overlap PCR connects the 5′ homology arm and orf+pa of GFP
[0...
Embodiment 2
[0300] Embodiment 2 experimental result and analysis
[0301] (1) Construction of plasmid CD144N-GFP (see results in figure 1 )
[0302] Using primers CD144N5F2 and CD144N5Rovr to amplify the 5'homologous arm LHA of 831bp; primers CD144N3F and CD144N3R1 amplified the 3'homologous arm SHA of 568bp; amplified GFP by primers CD144N-GFP-ovF and CD144N-GFP-R fragment. LHA and GFP were connected by overlapping primer extension method to form LHA-GFP. SHA and LHA-GFP were cut with PstI and NheI respectively and ligated to form an intermediate vector. The intermediate vector and F8-22-PGK-Neo were respectively digested with NheI enzyme Cut and connect PGK-Neo into the intermediate vector to form the final targeting vector CD144N-GFP.
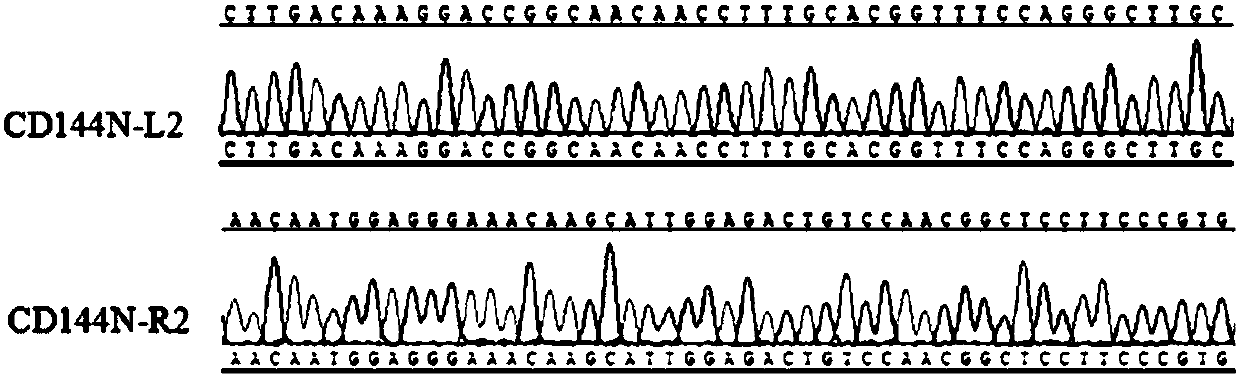
[0303] (2) Construction of plasmid TALEN (results see figure 2 )
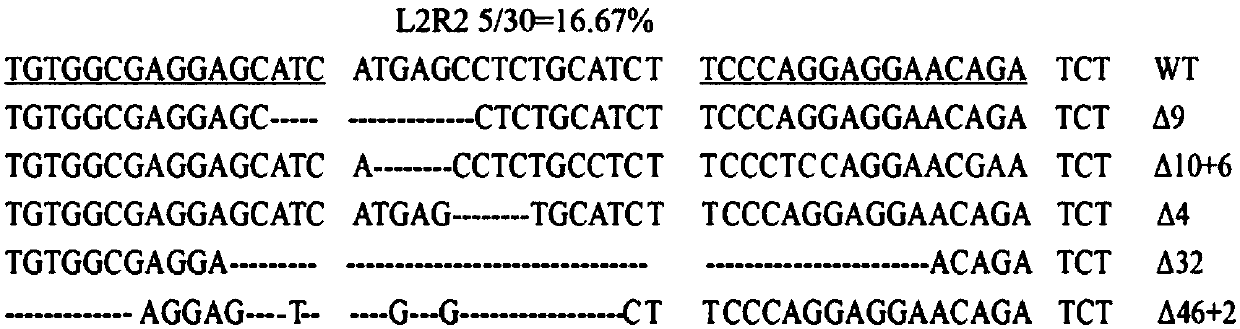
[0304] The TALENs targeting the N-terminus of the CD144 gene, CD144N-L2 and CD144N-R2, were constructed using the "REAL Assembly TALEN Kit" system established by the KeithJoung Lab...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com