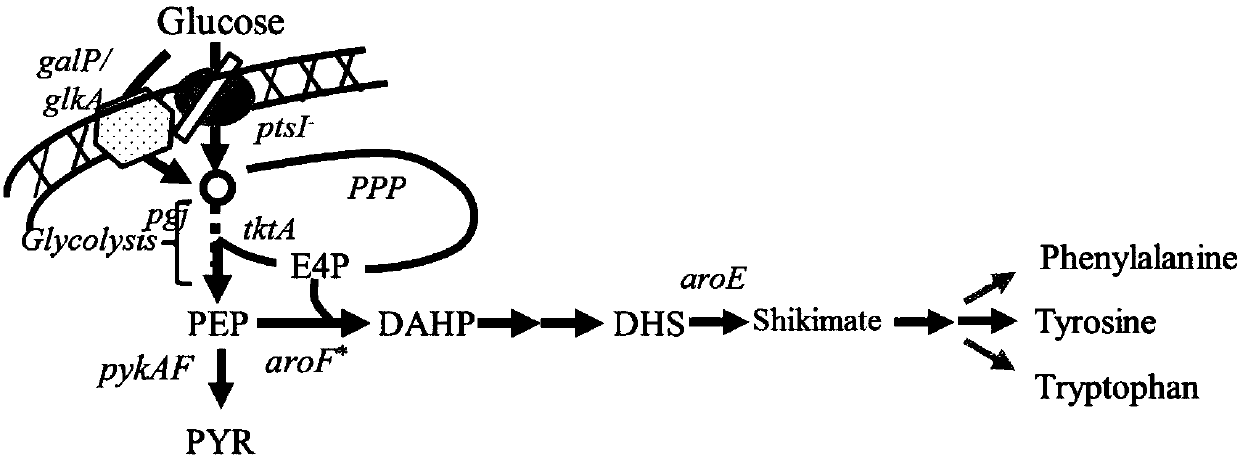
Escherichia coli recombinant bacterial strain for producing 3-dehydroshikimic acid as well as establishment method and application thereof
A technology of dehydroshikimic acid and recombinant strains, applied in the biological field, can solve problems affecting industrial applications, genetic instability, and production that cannot meet market demand, and achieve the effect of reducing production costs and medium costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0311] The construction of embodiment 1 escherichia coli recombinant strain WJ004
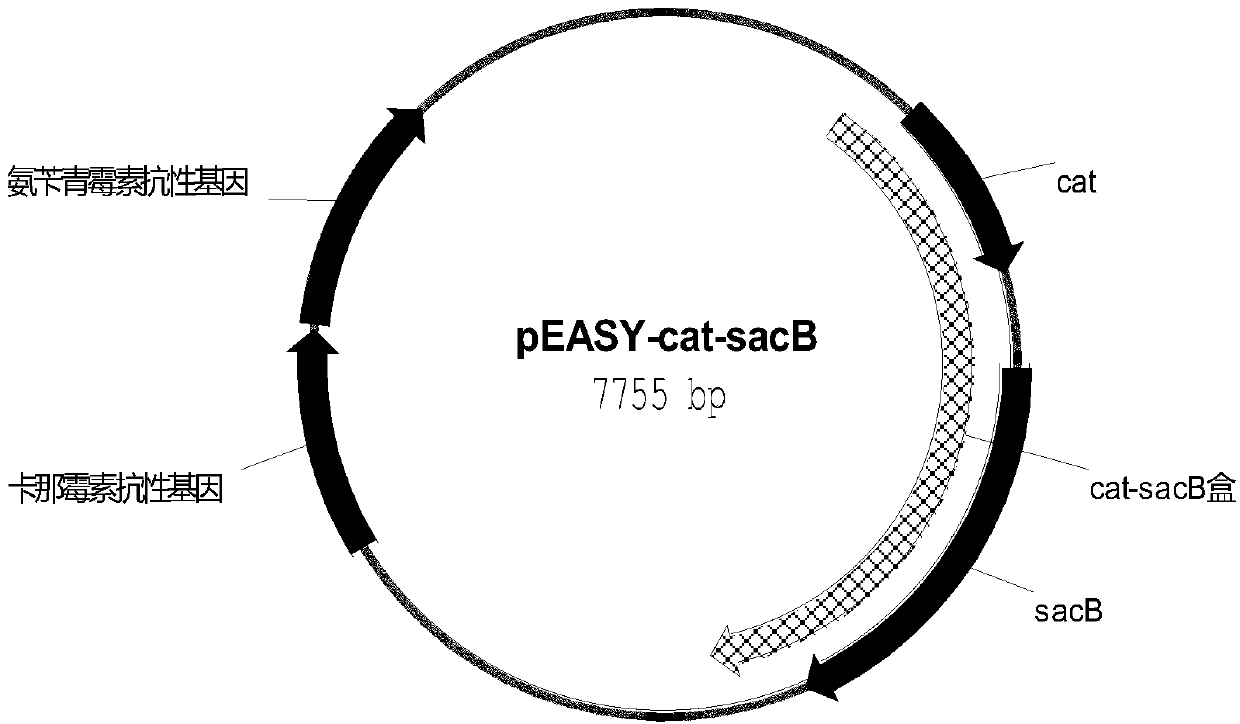
[0312] With the plasmid pEASY-cat-sacB (such as figure 2 (shown) is a template, and primer 41aroE1-up / aroE1-down is used to amplify the fragment aroE1 of homologous recombination in the first step. The sequence of primer 41 is:
[0313] aroE1-up (forward primer): GATGCCCTGACGGGTGAACTGTTTCGACAGGGGTAACATAGTGACGGAAGATCACTTC
[0314] aroE1-down (reverse primer): CTGTGGGCTATCGGATTACCAAAAACAGCATAGGTTTCCAATCAAAGGGAAAACTGTCC
[0315] Amplification system: 5×TransStart TM FastPfu Buffer 10 μL, dNTPs (2.5 mmol / L each dNTP) 4 μL, DNA template 1 μL (20-50ng), forward primer (10 μmol / L) 2 μL, reverse primer (10 μmol / L) 2 μL, 100% DMSO 1 μL, TransStart TMFastPfu DNA Polymerase (2.5U / μL) 1 μL, deionized water 29 μL, total volume 50 μL.
[0316] The amplification conditions are: 94°C pre-denaturation for 5 minutes (1 cycle); 95°C denaturation for 20 seconds, 55°C annealing for 30 seconds, 72°C extensio...
Embodiment 2
[0335] The construction of embodiment 2 escherichia coli recombinant strain WJ006
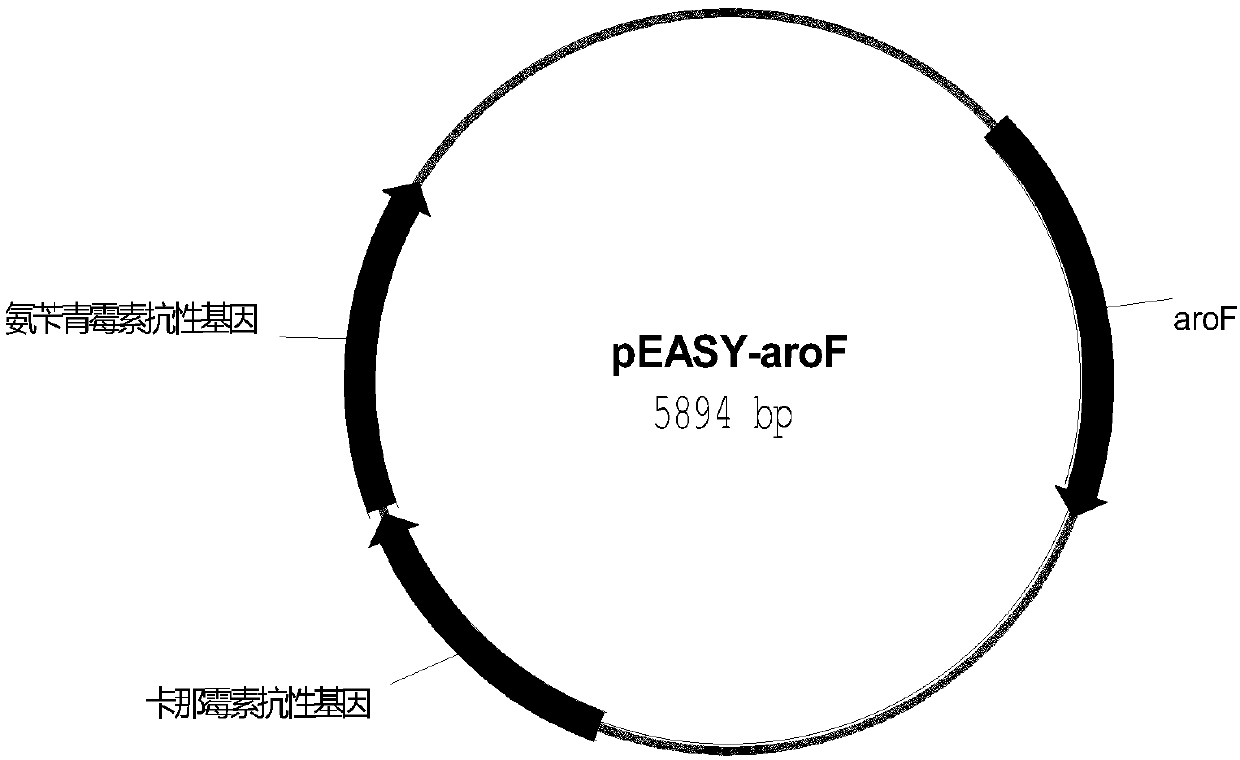
[0336] Using Escherichia coli DSM1576 genomic DNA as a template, the aroF gene was amplified using primer 61 aroF-F / aroF-R. The sequence of primer 61 is:
[0337] aroF-F (forward primer): ATGCAAAAAGACGCGCTGAA
[0338] aroF-R (reverse primer): TTAAGCCACGCGAGCCGTCAG
[0339] Amplification system: 5×TransStart TM Taq Buffer 5μL, dNTPs (2.5mmol / L each dNTP) 2μL, DNA template 1μL (20-50ng), forward primer (10μmol / L) 1μL, reverse primer (10μmol / L) 1μL, 100% DMSO 1μL, TransStart TM FastPfu DNA Polymerase (2.5U / μL) 1 μL, deionized water 29 μL, total volume 50 μL. Amplification conditions were pre-denaturation at 94°C for 5 minutes (1 cycle); denaturation at 95°C for 20 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 1 minute (30 cycles); extension at 72°C for 5 minutes (1 cycle).
[0340] The amplified aroF gene fragment was cloned into the pEASY-Blunt vector (purchased from Be...
Embodiment 3
[0384] Example 3 Construction of Escherichia coli recombinant strain WJ012
[0385] With the plasmid pEASY-cat-sacB (as shown in SEQ ID NO:7) containing chloramphenicol resistance gene cat and fructan sucrose transferase gene sacB figure 2 (shown) is a template, and primer 121tktA1-up / tktA1-down is used to amplify the fragment tktA1 of homologous recombination in the first step. The sequence of primer 121 is:
[0386] tktA1-up (forward primer): GCCCAAAACGCGCTGTCGTCAAGTCGTTAAGGGCGTGCCCTTCATCATGTGACGGAAGATCACTTC
[0387] tktA1-down (reverse primer): CATGCTCAGCGCACGAATAGCATTGGCAAGCTCTTTACGTGAGGACATATCAAAGGGAAAACTGTCC
[0388] Amplification system: 5×TransStart TM FastPfu Buffer 10 μL, dNTPs (2.5 mmol / L each dNTP) 4 μL, DNA template 1 μL (20-50ng), forward primer (10 μmol / L) 2 μL, reverse primer (10 μmol / L) 2 μL, 100% DMSO 1 μL, TransStart TM FastPfu DNA Polymerase (2.5U / μL) 1 μL, deionized water 29 μL, total volume 50 μL. Amplification conditions were pre-denaturation at...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com