Radix arnebiae polysaccharides and application thereof to preparation of anti-complement drugs
A technology of comfrey polysaccharide and application, applied in the field of natural homogeneous polysaccharide, preparation of anti-complement drugs, and polysaccharide, can solve the problems such as separation, preparation and activity report of homogeneous polysaccharide with anti-complement activity that have not yet been seen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1 Preparation of comfrey polysaccharides APS-1 and APS-2
[0022] 20Kg of comfrey is crushed, extracted by cold immersion in 95% ethanol, filtered, extracted 3 times with hot water, filtered, combined extracts, concentrated, centrifuged, added 4 times the volume of 95% ethanol to the supernatant, let stand, Centrifuge to remove the supernatant, redissolve the precipitate in water, recover under reduced pressure, and remove ethanol; then use trichloroacetic acid to remove free protein from the reconstituted solution, centrifuge, adjust the supernatant to neutral, dialyze, concentrate, and freeze-dry to obtain crude polysaccharide. Crude polysaccharide 0.5 g was dissolved in distilled water, centrifuged, and the supernatant was divided into DEAE-cellulose columns (Cl -1 type, 30×2.5 cm) chromatography for preliminary separation. Elute with distilled water, 0.4, 0.8, 1.2 and 2.0 mol / L NaCl solutions, the elution volume is greater than 2 times the column volume (a...
Embodiment 2
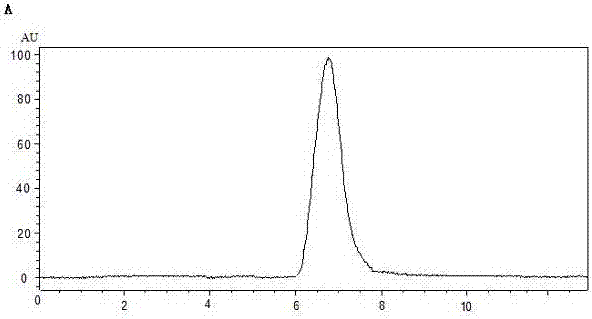
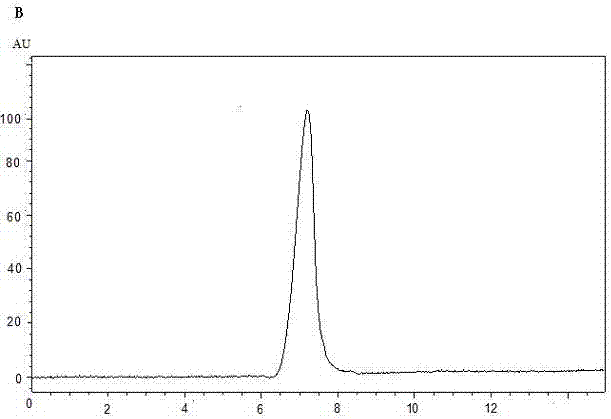
[0024] Example 2 Structural characterization of comfrey polysaccharides (APS-1 and APS-2)
[0025] (1) Molecular weight determination
[0026] APS-1 and APS-2 were compared with standard T-series dextran, and the results showed that the molecular weights of APS-1 and APS-2 were both less than 2×106 Da, but greater than 1×106 Da.
[0027] (2) Elemental analysis results
[0028] The elemental analysis results of APS-1 are: C, 42.35%; H, 6.59%; N, 1.84%.
[0029] APS-2 element analysis results are: C, 30.96%; H, 4.46%; N, 1.50%.
[0030] (3) Specific rotation
[0031] APS-1 specific curl: [ α ] D 25 =+14.2 ( c 0.318, H 2 O).
[0032] APS-2 specific curl: [ α ] D 25 = +24.7 ( c 0.205, H 2 O).
[0033] (4) Determination of total sugar, uronic acid, protein and sulfate content
[0034] The total sugar content of APS-1 determined by sulfuric acid-phenol method was 95.36%; the total sugar content of APS-2 was 95.81%.
[0035] The content of uronic acid in APS-1 was ...
Embodiment 3
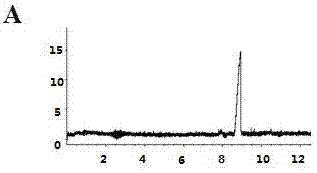
[0046] Example 3 Anti-complement classical pathway test in vitro
[0047] Take 0.1ml of complement (guinea pig serum), add BBS to prepare a 1:5 solution, and double-dilute with BBS to 1:10, 1:20, 1:40, 1:80, 1:160, 1:320 and 1: 640 solution. Dissolve 0.1 ml of 1:1000 hemolysin, each concentration of complement and 2% SRBC in 0.3 ml BBS, mix well, put in a low-temperature high-speed centrifuge at 37 ºC for 30 min, and centrifuge at 5000 rpm and 4 ºC for 10 min. Take 0.2 ml of the supernatant from each tube in a 96-well plate, and measure the absorbance at 405 nm. A full hemolysis group (0.1 ml 2% SRBC dissolved in 0.5 ml triple distilled water) was also set up in the experiment. The absorbance of three-distilled water lysed blood vessels was used as the standard of total hemolysis, and the hemolysis rate was calculated. Taking the dilution of complement as the X-axis, the percentage of hemolysis caused by each dilution of complement is plotted as the Y-axis. The lowest com...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com