Construction method of recombinant bacillus subtilis of high-yield pullulanase
A technology of Bacillus subtilis and pullulanase, which is applied in the field of construction of recombinant Bacillus subtilis and can solve problems such as no resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
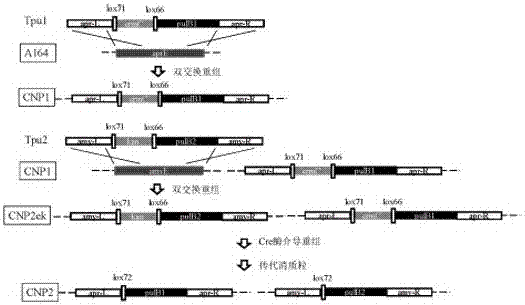
[0388] Embodiment 1: Construction of recombinant Bacillus subtilis CNP2
[0389] 1. Obtaining integrated DNA
[0390] There are two operons expressed by pullulanase, the first pulB1 Including artificial tandem promoter Pamy-cryIII, codon-optimized signal peptide sequence SPamyL secreted by amylase, and codon-optimized pullulanase coding gene pulB , pulB The sequence is shown in SEQ ID NO. 3, pulB1 The sequence is shown in SEQ ID NO. 10; the second pulB2 Including artificial tandem promoter PxylA-cryIII, codon-optimized chitinase secreted signal peptide sequence SPchit and codon-optimized pullulanase coding gene pulB , pulB2 See SEQ ID NO. 11 for the sequence.
[0391] for apr E Gene expression cassette of knockout recombination arm and erythromycin resistance fragment apr-erm-apr As shown in SEQ ID NO. 8, for amyE Gene expression cassette of knockout recombination arm and kanamycin resistance fragment amy-kan-amy As shown in SEQID NO. 9.
[0392] Bundle ...
Embodiment 2
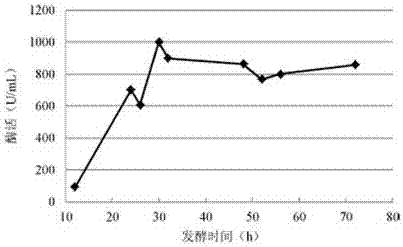
[0417] Embodiment 2: the fermentative production enzyme of recombinant Bacillus subtilis CNP2
[0418]Use recombinant Bacillus subtilis CNP2 as the production strain for shake flask fermentation: pick up recombinant Bacillus subtilis CNP2 with an inoculation loop and inoculate it into a 200 mL baffle bottle containing 25 mL liquid medium, 37°C, 200 r / min Fermentation culture is carried out, wherein the main components of the medium are corn steep liquor, maltose syrup, glucose, yeast powder and inorganic salt ions. After 20 hours of fermentation, samples were taken to measure the enzyme activity. The method for the determination of pullulanase activity was as follows: take 1.0 mL of acetic acid-sodium acetate buffer solution with pH=4.5 to prepare enzyme samples or diluted samples (the control group was samples inactivated in a boiling water bath), Add it to 1.0 mL of 0.5% pullulan solution, bathe in a water bath at 60°C for 30 min, then add 3.0 mL of 3,5-dinitrosalicylic acid...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com