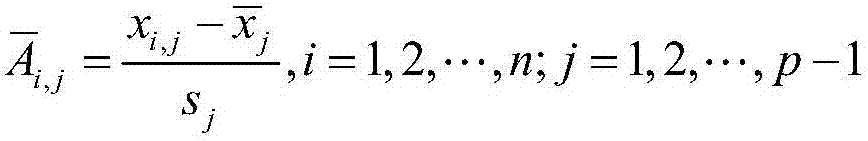Method for identifying cancer biomarkers
A biomarker and cancer technology, applied in the field of cancer biomarker identification, can solve the problems of poor promotion performance and achieve the effect of reducing bias and good promotion performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
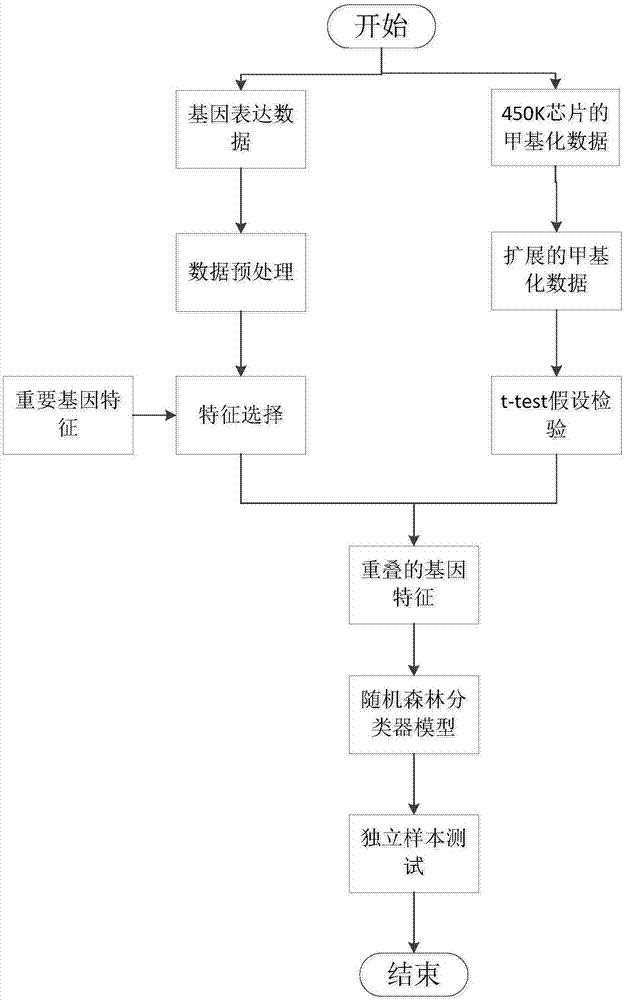
[0031] figure 1 It is a flowchart of a method for identifying cancer biomarkers in the present invention.
[0032] In this example, if figure 1 As shown, a method for identifying cancer biomarkers of the present invention comprises the following steps:
[0033] S1. Obtain the gene expression data and DNA methylation data of any cancer, as well as the known important genes corresponding to the cancer;
[0034] In this embodiment, the thyroid cancer THCA (thyroid carcinoma) is obtained from the cancer genome public database TCGA as an example to illustrate, and the corresponding DNA methylation data of the 450K chip, as well as the important THCA-related data reported in the literature Gene. Among them, the gene expression data of thyroid cancer THCA has 572 samples and 20503 gene features. The DNA methylation data of the 450K chip has 484 samples and 401833 site features.
[0035] S2, assuming that the gene expression data is a matrix of n×p, n is the number of rows of the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com