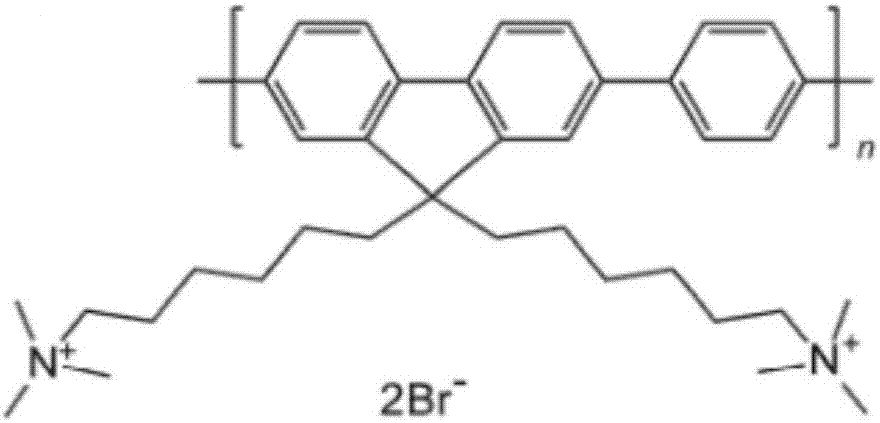
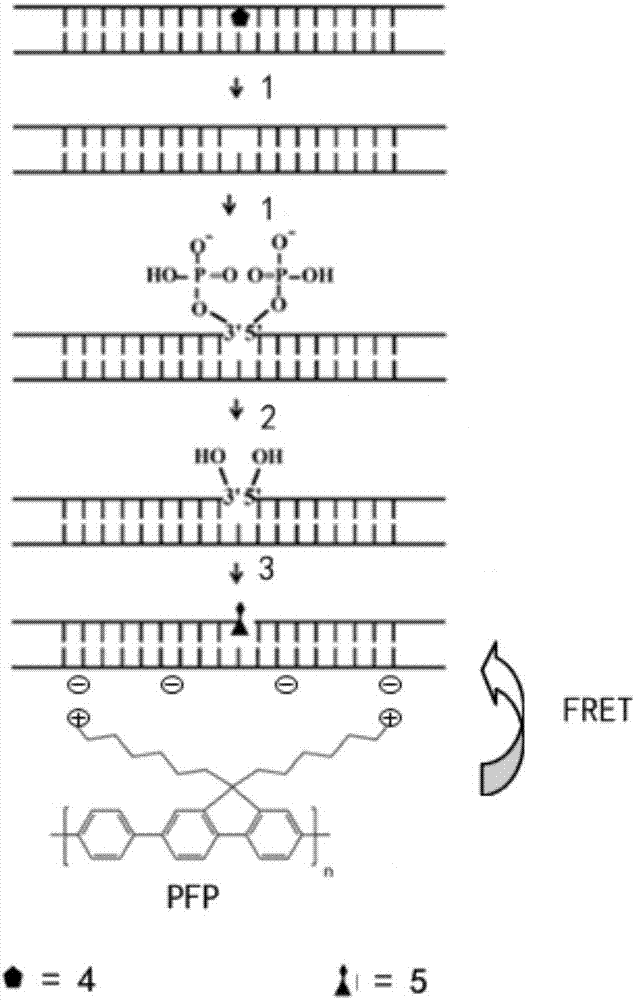
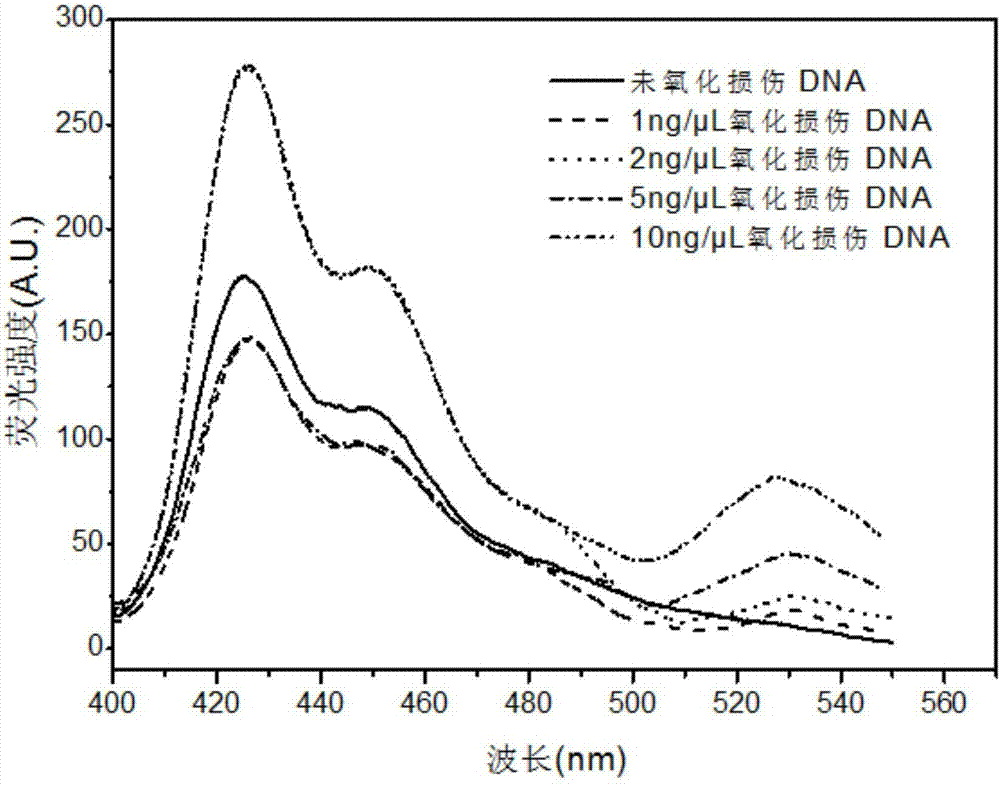
Oxidative damage DNA detection method
A technology of oxidative damage and buffer solution, which is applied in the fields of biochemical equipment and methods, and the determination/inspection of microorganisms. Achieve the effect of low detection limit, high sensitivity and easy promotion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0100] Preferably, the oxidative damage DNA standard sample is prepared by Fenton reaction, and its preparation method comprises the following steps:
[0101] (a) Dilute the extracted genomic DNA samples to different concentrations, and perform ultraviolet measurement to obtain the relationship between absorbance and DNA concentration;
[0102] (b) Take 10 μg of DNA samples with different concentrations and add 50 μL of 80 μM FeCl 2 , 40 μL 0.5 mM H 2 o 2 , 100 μL Tris-acetate buffer solution, reacted at 37°C for 4 hours in the dark; after the reaction, take it out quickly, add 20 μL 3M sodium acetate solution and 500 μL absolute ethanol, and react at -20°C for 10 minutes; centrifuge the solution to precipitate Wash the DNA pellet with 70% ethanol; dry the DNA pellet and redissolve it with 50 μL of water;
[0103] (c) performing ultraviolet measurement on the solution obtained in step (b), obtaining the DNA concentration in the solution according to the relationship between...
Embodiment approach
[0122] As a preferred embodiment, replace step (a') with the following steps:
[0123] Take 5 μg of oxidative damage DNA standard sample or 25 μL of detection sample, add 0.5 μL 10 U / μL formamide pyrimidine-DNA glycosylase, 5 μL NEB buffer 1 and 20 μL water, react at 37 °C for 90 minutes, and stop the reaction at 75 °C for 10 minutes minute;
[0124]The NEB buffer 1 comprises 10mM Bis-Tris-Propane-HCl, 10mM MgCl 2 and 1 mM DTT, pH 7.0 ± 0.5.
[0125] In this preferred technical scheme, step (a') can be performed not only using endonuclease VIII, but also using FPG protein. Endonuclease VIII mainly recognizes oxidized pyrimidine bases, and FPG protein mainly recognizes oxidized purine bases. Oxidative damaged DNA is transformed into DNA containing 3',5' phosphate group nucleotide gaps.
[0126] In order to further understand the present invention, the present invention will be further explained and described below in conjunction with specific examples.
[0127] The instrume...
Embodiment 1
[0129] Example 1 Effect of different concentrations of oxidatively damaged DNA on fluorescence detection signal
[0130] (a) The preparation method of oxidatively damaged DNA standard samples at different concentrations is as follows:
[0131] Dilute the extracted genomic DNA samples to different concentrations, and use a NanoDrop 2000 ultra-micro spectrophotometer to measure at the maximum absorption wavelength of 260nm to obtain the relationship between absorbance and DNA concentration;
[0132] Take 10 μg of DNA samples with different concentrations and add 50 μL of 80 μM FeCl 2 , 40 μL 0.5 mM H 2 o 2 , 100 μL Tris-acetate buffer solution (Tris-acetate buffer solution is a 2-fold concentrated buffer solution, which contains 20 mM Tris and 13 mM acetic acid, pH=7.3), reacted at 37 ° C for 4 hours in the dark; Take it out, add 20 μL of 3M sodium acetate solution (pH=5.2) and 500 μL of absolute ethanol (4°C), react at -20°C for 10 minutes, then take out the solution and cen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com