Corn ZmNST3 gene and application
A gene and corn technology, applied in the field of corn ZmNST3 gene and its application, to achieve the effect of increasing accumulation and facilitating development and utilization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 Cloning of ZmNST3
[0029] Total RNA was extracted from maize seedlings at the V3 stage, cDNA was synthesized by reverse transcription using the cDNA Synthesis Kit of Promega Company, and amplified with specific primers to obtain the ZmNST3 CDS sequence, with a full length of 1104bp (SEQ ID No.1). The nucleotide sequence of the primer pair is:
[0030] Upstream primer: 5'-TGATGAGCATCTCGGTGAACGG-3',
[0031] Downstream primer: 5'-GACTAGGCGTTGTTCATGGCCAA-3'.
Embodiment 2
[0032] Embodiment 2 is used for the construction of the expression vector expressing ZmNST3 gene in Arabidopsis and Agrobacterium transformation
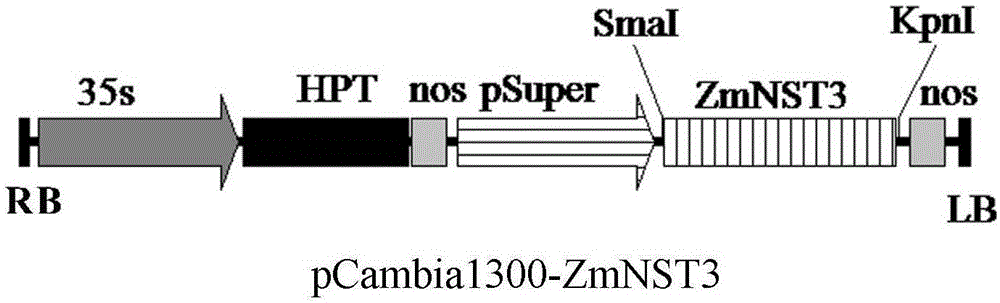
[0033] The expression cassette is obtained from the Super promoter of Agrobacterium tumefaciens (that is, the mas promoter is fused with 3 ocs enhancers and 1 mas enhancer, see Leisner SM, Gelvin SB (1988) Structure of the octopinesynthse upstream activator SEQuence. Proc Natl Acad Sci USA 85, 2553-2557) and the 3' transcription termination region from the nopaline synthase (nos) gene (Depicker A, Stachel S., Dhaese P, Zambryski P, Goodman HM (1982). Nopaline synthase: transcript mapping and DNASEQuence.J Mol.Appl Genet.1,561-573), the selection marker gene is hygromycin phosphotransferase gene (hpt). Its T-DNA region such as figure 1 shown.
[0034] Using the plasmid containing the ZmNST3 gene as a template, use the PCR method to obtain a ZmNST3 gene fragment with XbaI and SmaI restriction sites at both ends, connect it to the pM...
Embodiment 3
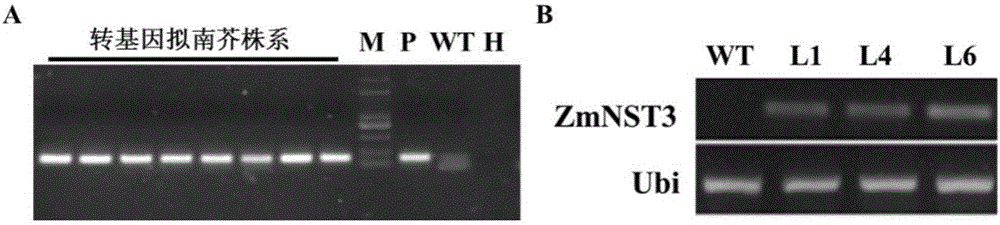
[0035] Example 3 Construction and Genetic Transformation of Expression Vectors for Expressing ZmNST3 in Maize
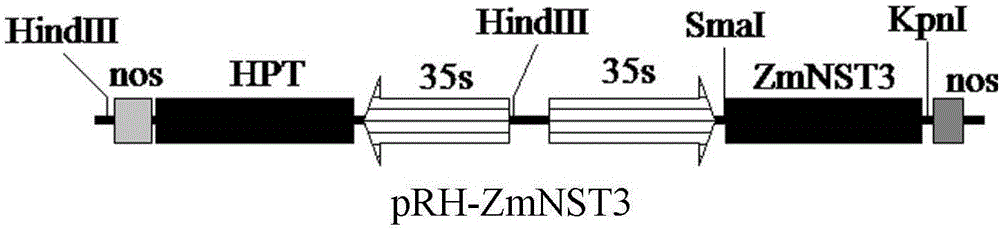
[0036] The expression cassette is composed of the CaMV35s promoter and the 3' transcription termination region from the nopaline synthase (nos) gene, and the selectable marker gene is the hygromycin phosphotransferase gene (hpt). Its expression box is as figure 2 shown.
[0037] Using the plasmid containing the ZmNST3 gene as a template, use the PCR method to obtain a ZmNST3 gene fragment with SmaI and KpnI restriction sites at both ends, connect it to the pMD19T vector, perform double digestion with SmaI and KpnI after the sequence is correct, and recover about 1100bp fragments. The expression vector pROK219 (containing the CaMV35s promoter) was double digested with SmaI and KpnI, and the large fragment of the vector was recovered. The vector and the target fragment were connected, and the E. coli competent cells were transformed to obtain the recombinant plasmi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com