Primer combination for identifying three medicinal snakes and application thereof
A primer combination, primer pair technology, applied in recombinant DNA technology, microbial determination/inspection, biochemical equipment and methods, etc., can solve the problems of long testing time, unfavorable application and promotion, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] Embodiment 1, primer design
[0093]Extensive sequence analysis of the genomes of various snake species yielded several primers for the identification of vipers, vipers, and vipers. Preliminary experiments were carried out on each primer to compare the performance of sensitivity and specificity, and finally three sets of primer pairs were obtained for the identification of vipers, black snakes and white snakes.
[0094] The primer pair (primer pair I) used to identify Vipers consists of primer F1 and primer F2 (5'→3'):
[0095] F1 (sequence 1 of the sequence listing): CTATACCTAATATTCGGCGCT;
[0096] F2 (sequence 2 of the sequence listing): CGGAGAGAGGTGGATAGACG;
[0097] The primer pair (primer pair II) used to identify black snakes consists of primer F3 and primer F4 (5'→3'):
[0098] F3 (sequence 3 of the sequence listing): CAGACATAGCCTTCCCACGC;
[0099] F4 (sequence 4 of the sequence listing): GGGGGGTATACTGTTCACCCA;
[0100] The primer pair (primer pair III) used...
Embodiment 2
[0104] Embodiment 2, identification method establishment
[0105] 1. Agarose gel electrophoresis
[0106] 1. Extract the genomic DNA of the sample to be tested.
[0107] 2. Using the genomic DNA obtained in step 1 as a template, use primer pair I, primer pair II and primer pair III in the primer combination prepared in Example 1 to perform PCR amplification respectively to obtain amplified products.
[0108] PCR amplification system (25 μL): 2.5 μL 10×buffer, 1.6 μL dNTP (2.5 mM), 0.4 μL primer F1, 0.4 μL primer F2, 0.4 μL primer F3, 0.4 μL primer F4, 0.4 μL primer F5, 0.4 μL μL primer F6, 0.2 μL SpeedStar HS Taq DNA polymerase, 2 μL template, 16.3 μL sterile double distilled water.
[0109] PCR reaction program: pre-denaturation: 95°C, 1min; denaturation: 95°C, 10s, annealing: 60°C, 10s, 30 cycles; 4°C storage.
[0110] 3. Perform 1.5% agarose gel electrophoresis on the amplified product obtained in step 2, and judge as follows according to the result:
[0111] If the amp...
Embodiment 3
[0121] Embodiment 3, identification method verification
[0122] Samples to be tested: 37 samples of snake medicinal materials of known species from different collection places shown in Table 1. The samples of Chinese herbal medicines in the table are all in compliance with the relevant regulations under the main text of the Chinese Pharmacopoeia (2015 edition) under each medicinal material. Through the identification, the real medicinal materials of each flavor are consistent with the names, and the quality meets the standards.
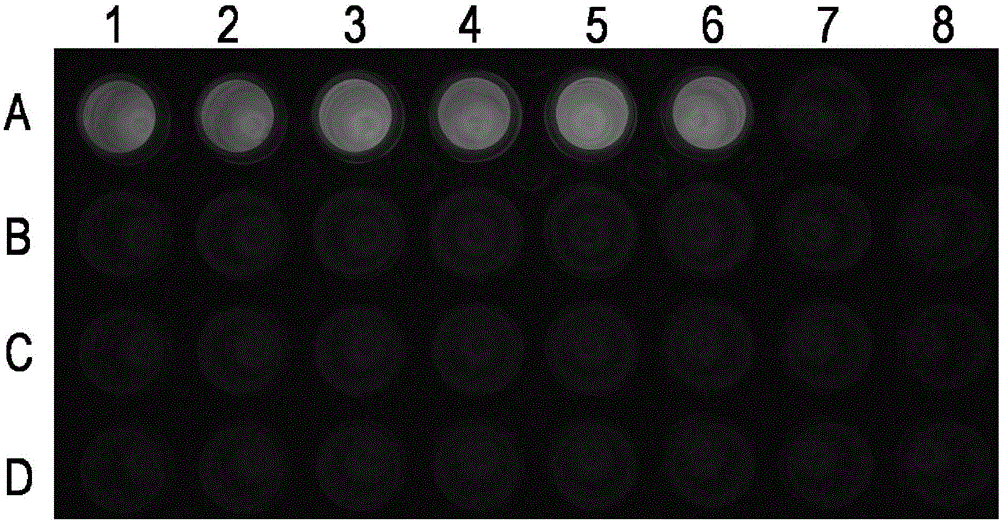
[0123] The samples to be tested were identified by the agarose gel electrophoresis method and the fluorescent staining method established in Example 2. In the PCR reaction system, the DNA content of the template was 20 ng.
[0124] Table 1 Sample source table
[0125]
[0126]
[0127]
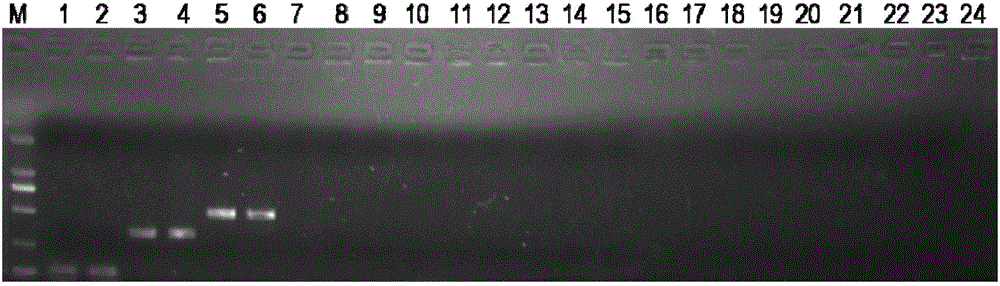
[0128] Some results of identification by agarose gel electrophoresis can be found in figure 1 . Will figure 1 The 124bp target bands in lane 1 and lane...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com