A highly efficient genome-wide chromatin conformation technique ehi-c
A chromatin, ehi-c technology, applied in recombinant DNA technology, combinatorial chemistry, microbial assay/inspection, etc., can solve the problems of increased cell culture cost, increased experimental error, low interaction resolution, etc. Cell requirements, savings in reagents, effect of simplified steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1. Preparation of cell eHi-C sequencing library and its application in sequencing (5-standard)
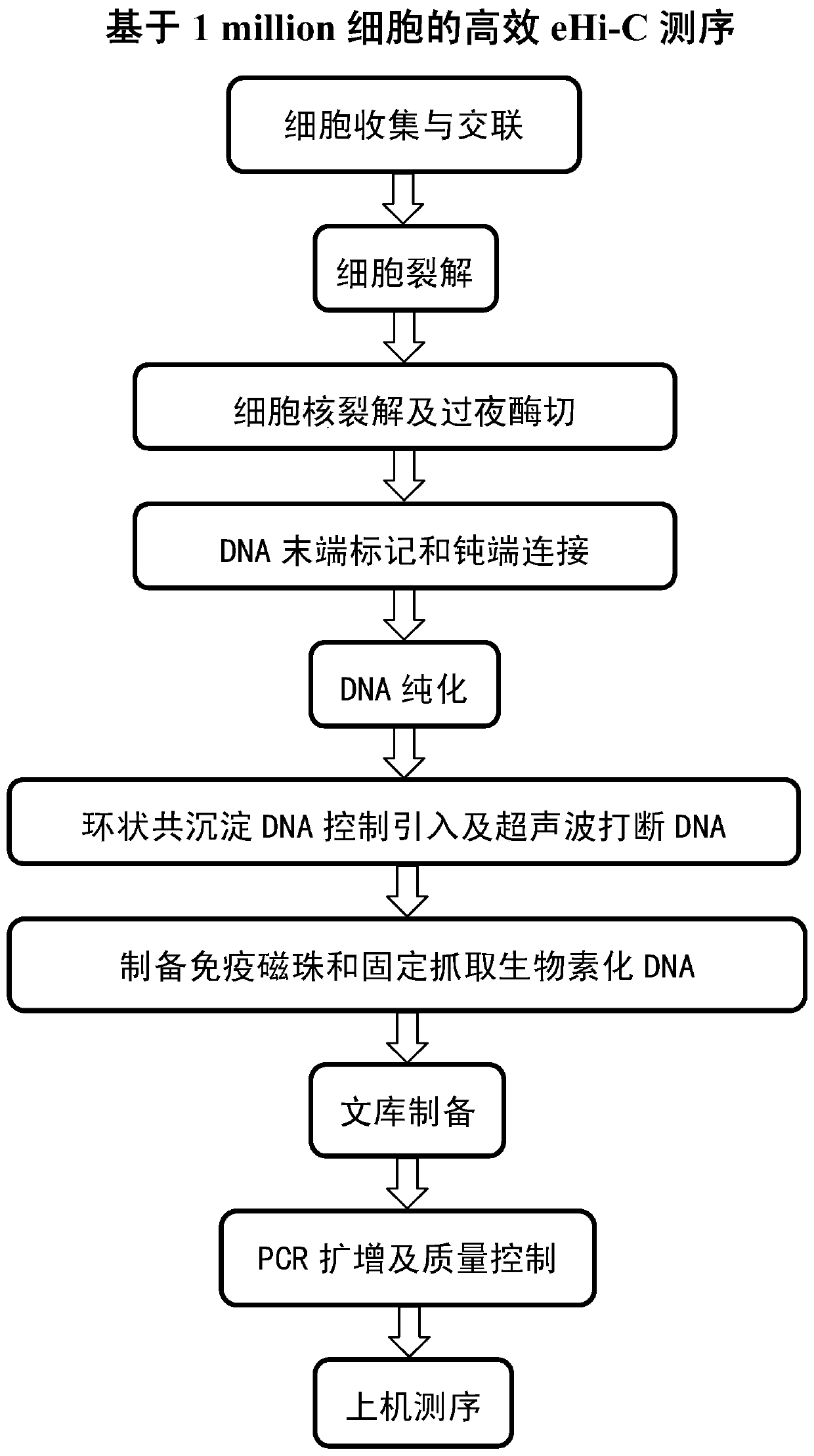
[0069] figure 1 It is a flowchart of main steps of the present invention.
[0070] (1) Lysis cells to prepare chromatin
[0071] 1. Cell collection and cross-linking fixation
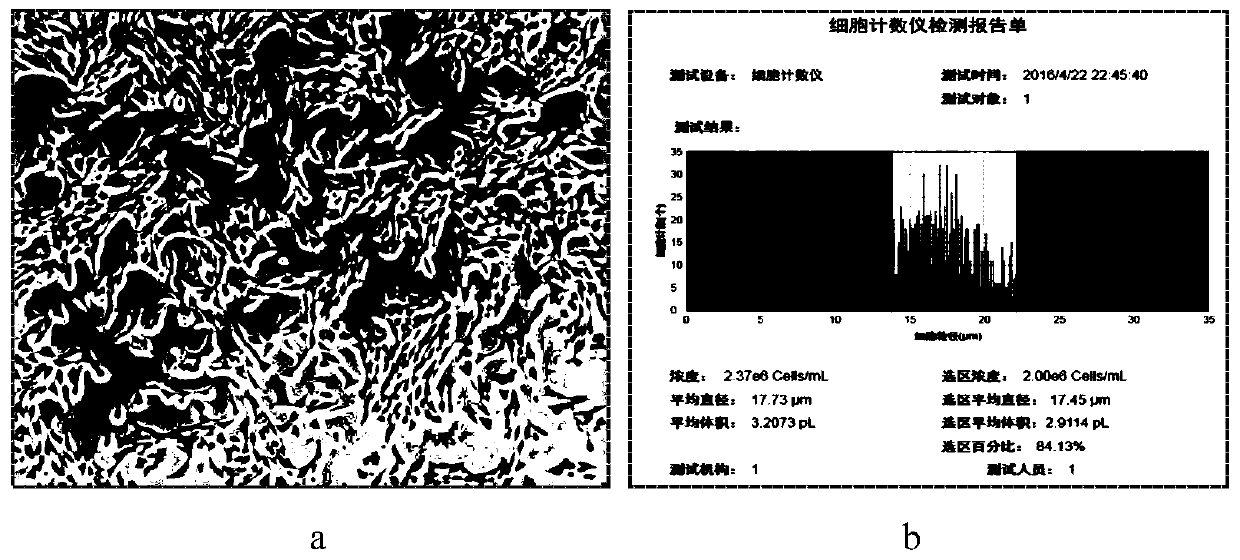
[0072] 1. When the cell confluence rate in the culture dish (10CM) reaches 70-80%, the C2C12 cells ( CRL-1772 TM ) counts about 2million, such as image 3 , a, cell state (magnification 100×); b, result of automatic cell counter), the volume of the culture solution was detected by pipetting;
[0073] 2. Add 37% formaldehyde aqueous solution (280 ul) by mass percentage to it to make the final concentration 1%, put the petri dish on a rotator and shake it at room temperature for 10 minutes (or incubate at 37° C. for 10 minutes);
[0074] 3. Add 2.5M glycine aqueous solution (0.89ml, so that the final concentration is 0.2M) to quench the formaldehyde, put it back into the rotator and shak...
Embodiment 2
[0150] Example 2, other methods of cell eHi-C sequencing
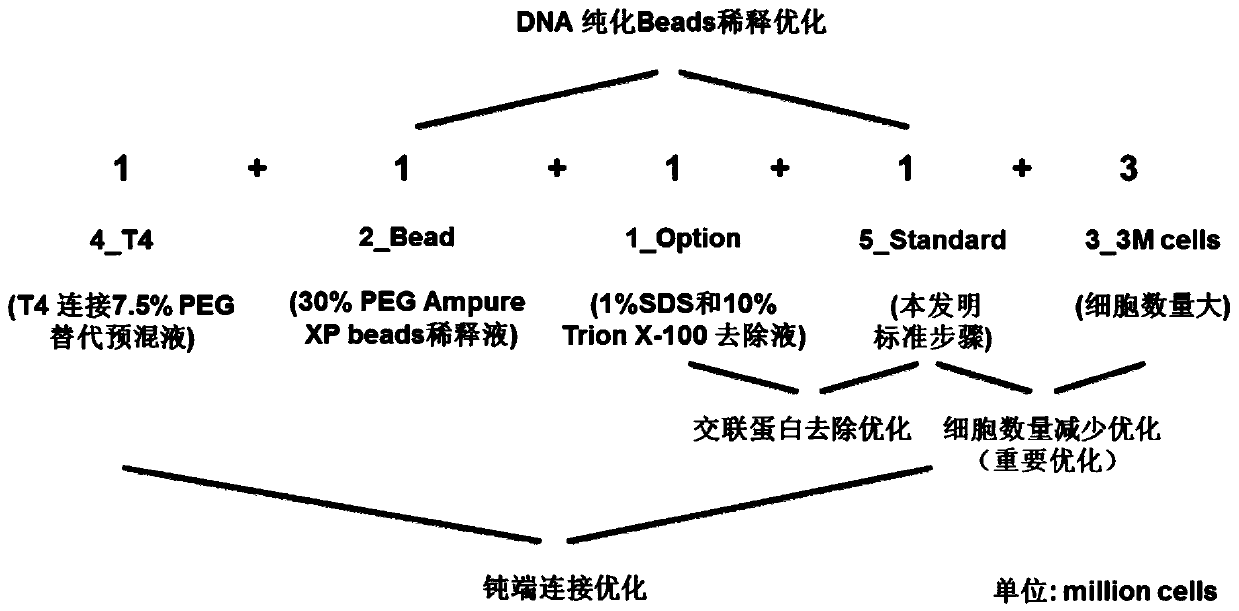
[0151] 1. Method ( figure 2 shown):
[0152] 1. 1-Option group: the method is basically the same as in Example 1, the only difference is (2), to obtain the 1-Option group eHi-C sequencing library:
[0153] Use 500ul of 1×NEBuffer 2 (NEB; #B7002S) to wash the chromatin aggregate obtained in (1) above, centrifuge at 20,000g for 15min at 4°C, and remove the supernatant. repeat. Break up the clumps again with a pipette tip, add 38ul 1×NEBuffer 2, resuspend and mix as completely as possible, and obtain the resuspended chromatin.
[0154] Add 3.8 ul of 1% SDS aqueous solution to the resuspended chromatin, so that the final concentration of SDS is 0.1%, mix well, and incubate at 65°C for 10 minutes to remove proteins that are not directly cross-linked with DNA; after incubation Immediately put the centrifuge tube on ice, add 4.4ul volume percentage of 10% Triton X-100 to quench SDS, mix carefully to avoid air bubbles; get...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com