A co-enrichment medium ses
A technology of enrichment medium and basic medium, which is applied in the field of agricultural product testing, can solve the problems that high-throughput detection methods are difficult to detect and cannot bypass the enrichment, so as to save quantity, avoid the interference of bacteria, and test samples wide range of effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
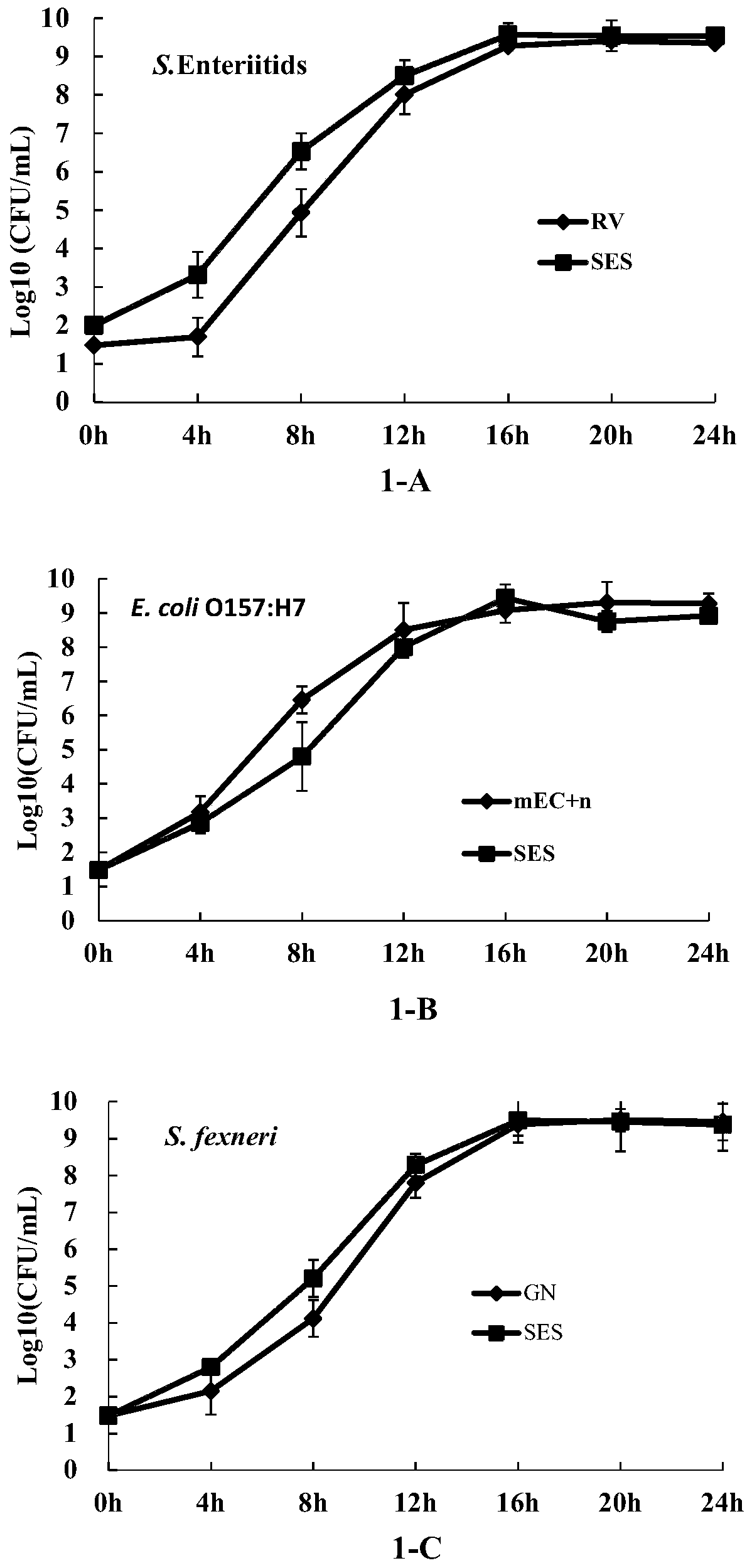
[0035] Example 1: SES monoenrichment effect identification
[0036] Preparation of SES medium: (1) Basal medium: 20 parts of tryptone, 1 part of glucose, 5 parts of NaCl, 4 parts of dipotassium hydrogen phosphate, 1.5 parts of potassium dihydrogen phosphate; (2) Inhibitor: No. 3 bile salt 1.2 parts, 20 parts of novobiocin sodium salt, 0.5 parts of lithium chloride, 0.1 parts of potassium tellurite; (3) distilled water, 1000 parts. Weigh tryptone, glucose, NaCl, dipotassium hydrogen phosphate, potassium dihydrogen phosphate, No. 3 bile salt, lithium chloride, add water and stir evenly, autoclave at 121°C, cool to room temperature, in the above mixture, no Add novobiocin sodium salt and potassium tellurite to the bacteria, and mix well to prepare the required co-enrichment medium (SES).
[0037] SES monoenrichment culture: take the overnight culture of Salmonella enteritidis (RV), Escherichia coli (mEC) O157:H7 and Shigella flexneri (GN) and inoculate 100ml of SES and their res...
Embodiment 2
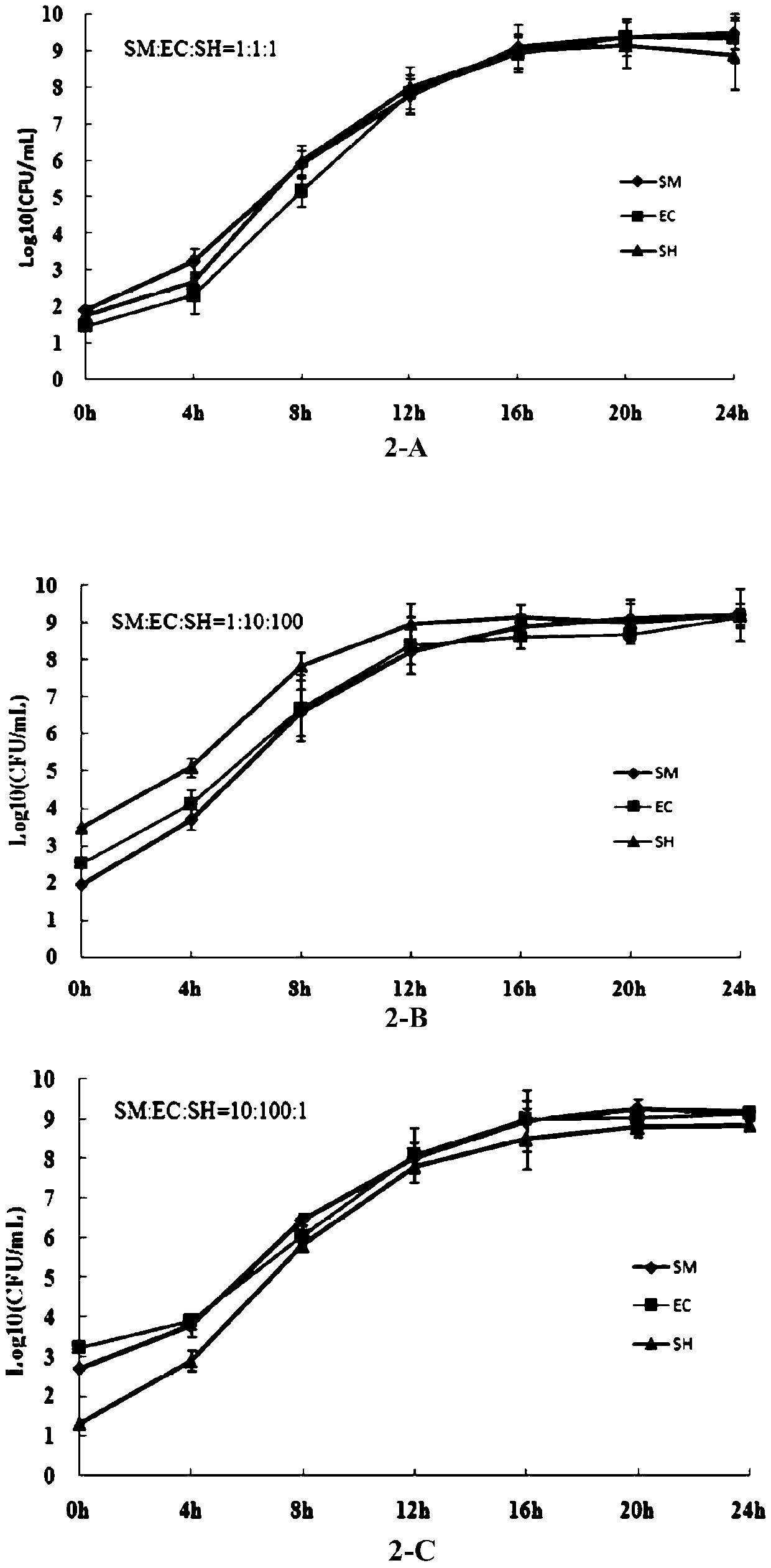
[0039] Example 2: Identification of SES compound bacterial enrichment effect
[0040] Preparation of SES medium: refer to Example 1.
[0041]SES compound enrichment culture: Three target bacteria, Salmonella enteritidis, Escherichia coli O157:H7 and Shigella flexneri, were inserted into SES at different concentration ratios. The proportions of the compound inoculation of the above three kinds of bacteria were as follows: 1:1:1, 1:10:100, 10:100:1 and 100:1:10. Cultivate at 36°C and 180rpm / min for 24h, and take samples every 4h for colony counting. The enumeration medium of each target bacteria was selected from its own selective plate: BS for Salmonella enteritidis, CT-SMAC for Escherichia coli O157:H7, and selective plate for Shigella flexneri.
[0042] SES compound bacterial enrichment effect identification: according to the colony counting results of each time period, the growth curve was drawn ( figure 2 ), comparing the growth ability of target bacteria with different ...
Embodiment 3
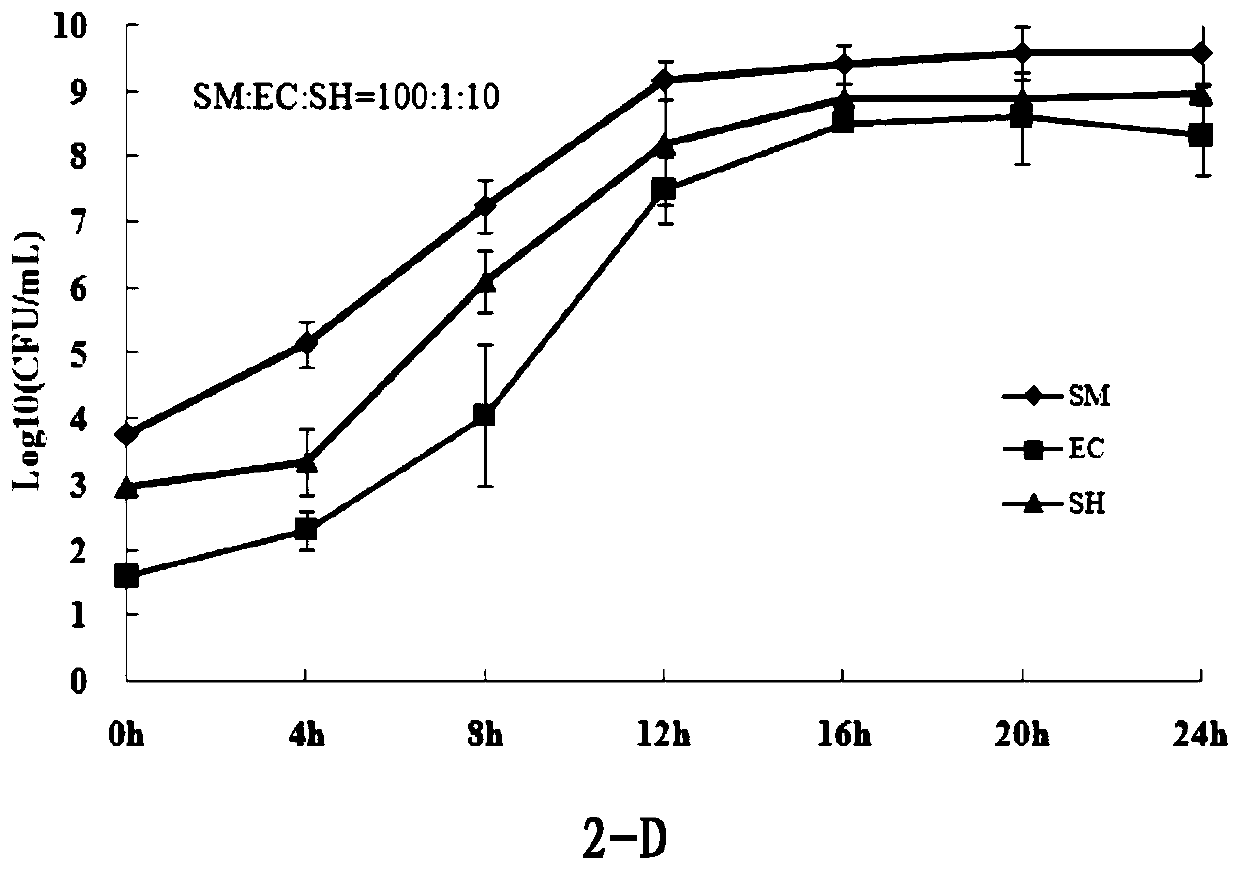
[0043] Example 3: Detection of 3 foodborne pathogens in frozen pork by SES combined with multiplex PCR
[0044] Preparation of SES medium: refer to Example 1.
[0045] Treatment of samples to be tested: Take 10 g of commercially available raw pork, soak the pork in alcohol, disinfect and air-dry, inoculate Salmonella Enteritidis, Escherichia coli O157:H7, and Shigella flexneri respectively at a final concentration of about 10 CFU / g, and air-dry to The bacterial solution was completely absorbed, and stored in a homogeneous bag at -20°C for 7 days.
[0046] SES enrichment culture and effect identification: put 10g of the artificially polluted pork sample in 90ml SES, and culture it at 36°C for 24h. From 12h, take the enrichment solution every 4h to count the viable bacteria of the above three target bacteria, and identify the SES. Bacteria effect ( image 3 ). The three target bacteria can still be enriched by SES after being frozen in pork at -20°C for 7 days, and the final ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com