Method for CRISPR/Cas9 targeted knockout of human ezrin gene enhancer key region and specific gRNA thereof
A key region, specific technology, applied in the field of gRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 gRNA design and vector construction targeting the key region of human ezrin gene enhancer
[0026] 1. Design of gRNA targeting the key region of enhancer of human ezrin gene and synthesis of oligonucleotide chain
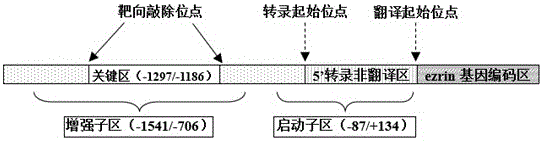
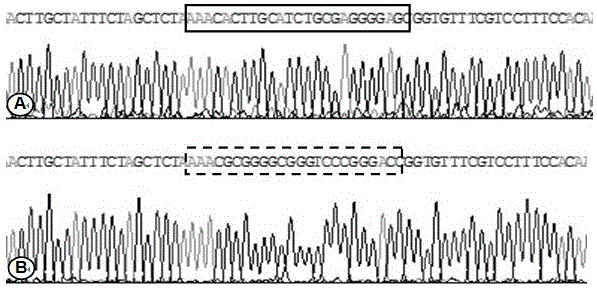
[0027] Find the human ezrin gene sequence from Genebank ( http: / / www.ncbi.nlm.nih.gov / gene / 7430 ), using the online software http: / / www.e-crisp.org / E-CRISP / The gRNA target sites were designed to be located upstream and downstream of the enhancer key region (–1297 / –1186) of the human ezrin gene, respectively. In the present invention, the DNA sequence corresponding to the gRNA is also called the gRNA sequence, which is the target site of the gRNA on the target gene. The oligonucleotide chain (Oligo DNA) corresponding to the gRNA is in accordance with the 5'-G(N) 20The PAM structure (protospacer adjacent motif) of NGG-3' is the design principle, and the sequence with a higher score is selected. If the first base at the 5' end of the forward ol...
Embodiment 2
[0036] Example 2 Cell culture and CRISPR / Cas9 recombinant plasmid transfection
[0037] Human esophageal cancer EC109 cells were grown adherently in DMEM medium containing 10% inactivated fetal bovine serum, digested with digestive solution containing 0.25% trypsin and 0.02% EDTA, and subcultured. Cells were seeded in 96-well cell culture plates, 100 μl per well. After 24 hours, the cells can be used for transfection when the confluence rate reaches 50%-60%. Co-transfection of plasmids pX459-sgRNA1 and pX459-sgRNA2 by liposome method, transfection steps refer to Lipofectamine TM 2000 Transfection Reagent instructions were performed.
Embodiment 3
[0038] Example 3 Targeted knockout detection of key regions of ezrin enhancer in human esophageal cancer cells
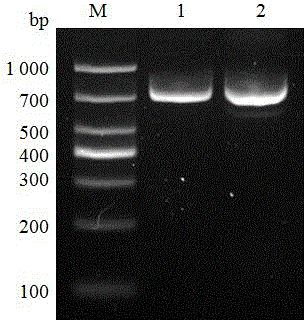
[0039] The recombinant plasmids pX459-sgRNA1 and pX459-sgRNA2 were co-transfected into esophageal cancer EC109 cells for 48 hours, and the cells were collected directly without resistance selection. Using cell genome DNA as a template, PCR amplifies the DNA sequence of the ezrin gene enhancer. The PCR primers are located upstream and downstream of the sequence to be knocked out, and the primer sequences are shown in SEQ ID NO.8 and SEQ ID NO.9.
[0040] The sequences of gRNA1 and gRNA2 are respectively located on both sides of the key region of the ezrin enhancer. It is estimated that the amplified fragment of non-mutated genomic DNA is 766 bp (ezrin gene -1543 / -778 sequence); the mutant genomic DNA is deleted by 147 bp (ezrin gene -1319 / -1173 sequence ), the amplified fragment is about 619bp in length. The results of agarose gel electrophoresis of PCR products ar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com