Preparation method and application of phi29 DNA polymerase
A polymerase and double-enzyme cleavage technology, applied in the field of phi29DNA polymerase preparation, can solve the problems of no disclosure of Phi29DNA polymerase preparation method, no disclosure of high-yield Phi29DNA polymerase preparation method, etc., to achieve low cost, stable quality, and distribution uniform effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
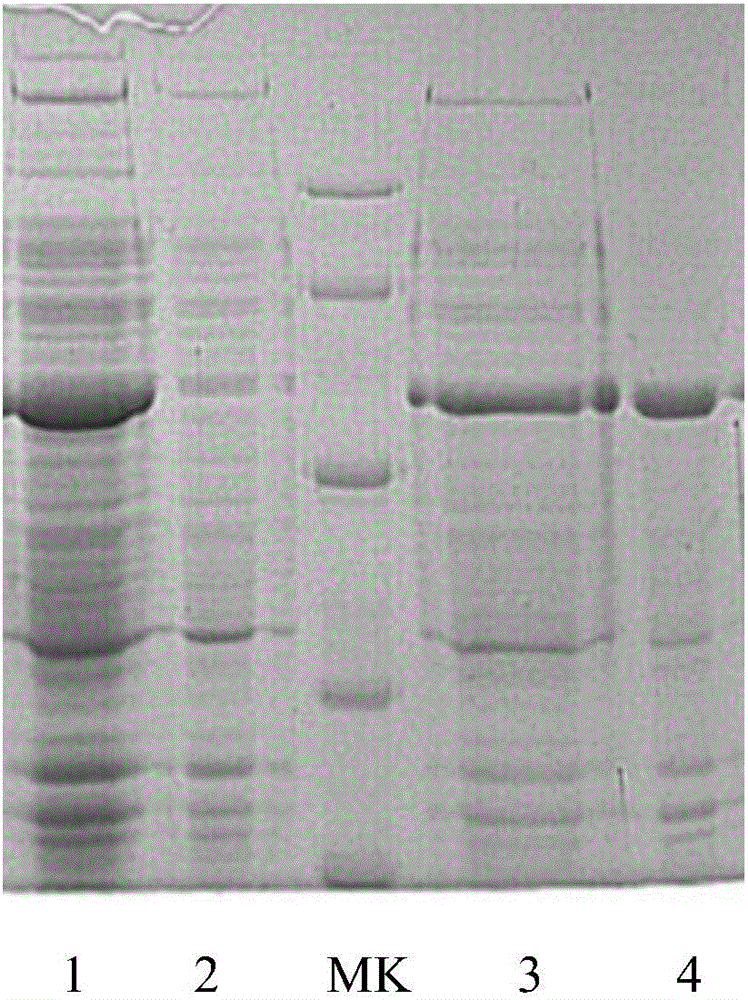
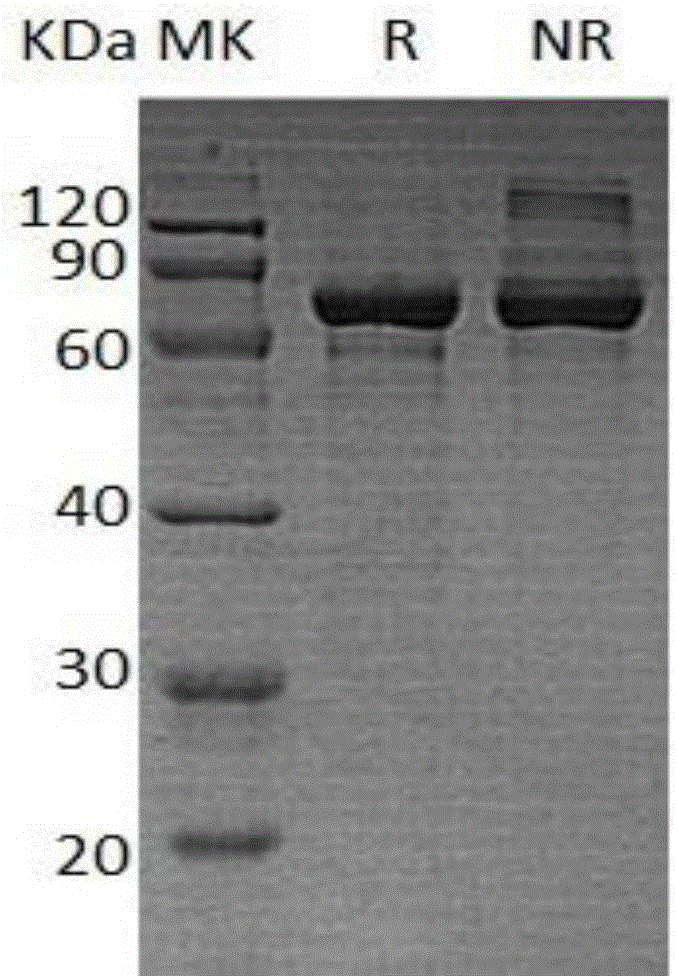
[0034] The preparation of embodiment 1.phi29DNA polymerase
[0035] 1. Construction of phi29 expression plasmid
[0036] The amino acid sequence of phi29 DNA polymerase is as shown in SEQ ID NO: 2. First, the gene sequence of phi29 DNA polymerase is synthesized from the whole gene, that is, the phi29 gene (SEQ ID NO: 1), and the synthetic phi29F (SEQ ID NO: 3:
[0037] CGCATATGATGAAGCACATGCCGCGCAAG) primers and phi29R (SEQIDNO:4:CGCTCGAGCTTGATGGTGAAGGTATC) primers were used for PCR amplification, and the PCR products were recovered and connected to the pET21a expression vector digested with NdeI and XhoI, transformed into Escherichia coli DH5α, and sequenced to obtain the correct clone plasmid.
[0038] 2. Expression of phi29 recombinant protein
[0039] The obtained correct cloning plasmid is transformed into the host bacteria, and the transformed monoclonal bacteria are inoculated in the test tube.
[0040] (1) Shake the monoclonal cells at 37°C for 4 hours, add 0.5mM IP...
Embodiment 2
[0066] Embodiment 2.Amplification of DNA Marker template plasmid
[0067] 1. Sample pre-denaturation: take the DH5α bacterial liquid containing the PUC19 plasmid, and refer to the following reaction system for thermal denaturation;
[0068] 2. Plasmid amplification: proceed according to the following reaction conditions;
[0069]
[0070] 3. Heat inactivation: 65°C, 10 minutes;
[0071] 4. Sequencing: Take 2 μl of the above reaction solution, add 8 μl of double distilled water, and perform direct sequencing.
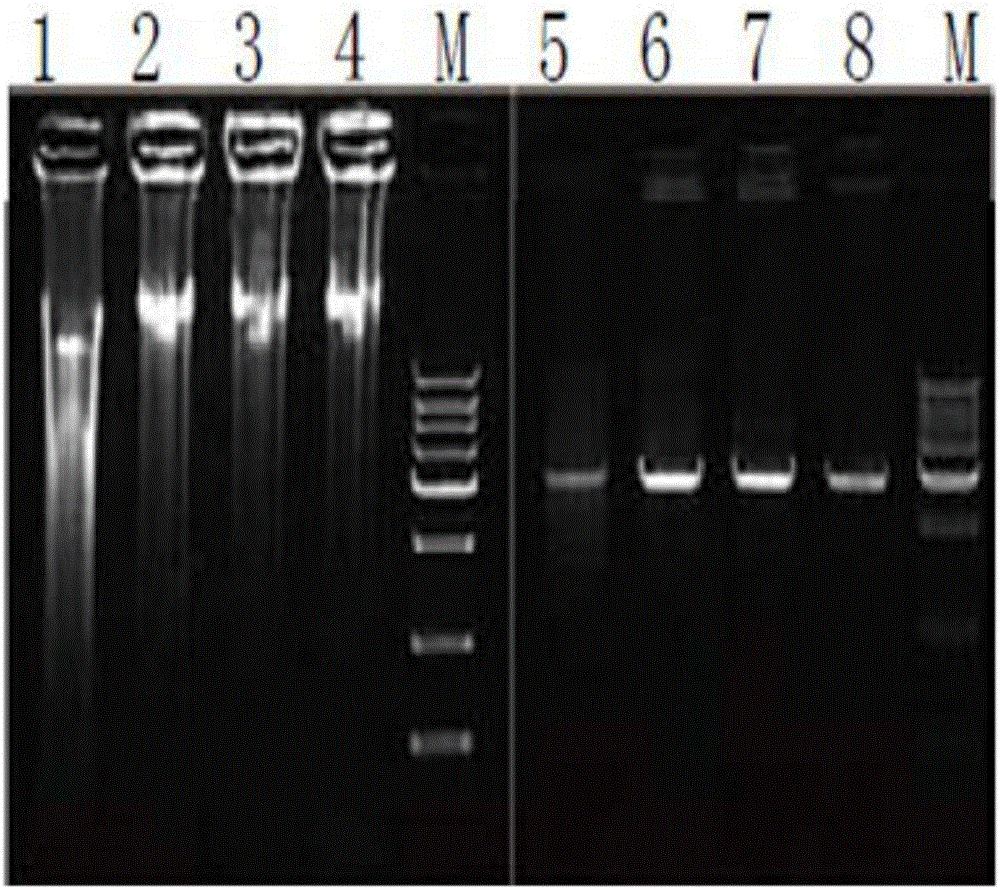
[0072] The result is as image 3 shown, from image 3 It can be seen that directly using the bacterial liquid as a template, a small amount of bacterial liquid (0.5 μl) can be amplified to obtain a large number of plasmid templates, which saves time and is easy to operate.
[0073] 5. Using the PUC19 plasmid obtained from the above amplification as a template, different primer pairs were designed by PCR method to amplify DNAs of 5kb, 3kb, 2kb, 1.5kb, 1kb, 750bp,...
Embodiment 3
[0074] Embodiment 3, the amplification of DNA Marker template plasmid
[0075] 1. Sample pre-denaturation: take the frozen DH5α bacterial liquid containing pPIC9k plasmid, and refer to the following reaction system for thermal denaturation;
[0076] 2. Plasmid amplification: proceed according to the following reaction conditions.
[0077]
[0078] 3. Heat inactivation: 65°C, 10 minutes.
[0079] 4. Sequencing: Take 2 μl of the above reaction solution, add 8 μl of double distilled water, and perform direct sequencing.
[0080] The result is as Figure 5 shown, from Figure 5 It can be seen that directly using the glycerol bacterial liquid as a template, a small amount of glycerol bacterial liquid (0.5 μl) can be amplified to obtain a large number of plasmid templates, which saves time and is easy to operate.
[0081] 5. Using the pPIC9k plasmid obtained by the above amplification as a template, different primer pairs were designed by PCR method to amplify DNA fragments...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com