Method for isolating nucleic acids
A nucleic acid and cell separation technology, applied in the field of nucleic acid separation to achieve the effect of simplifying procedures
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
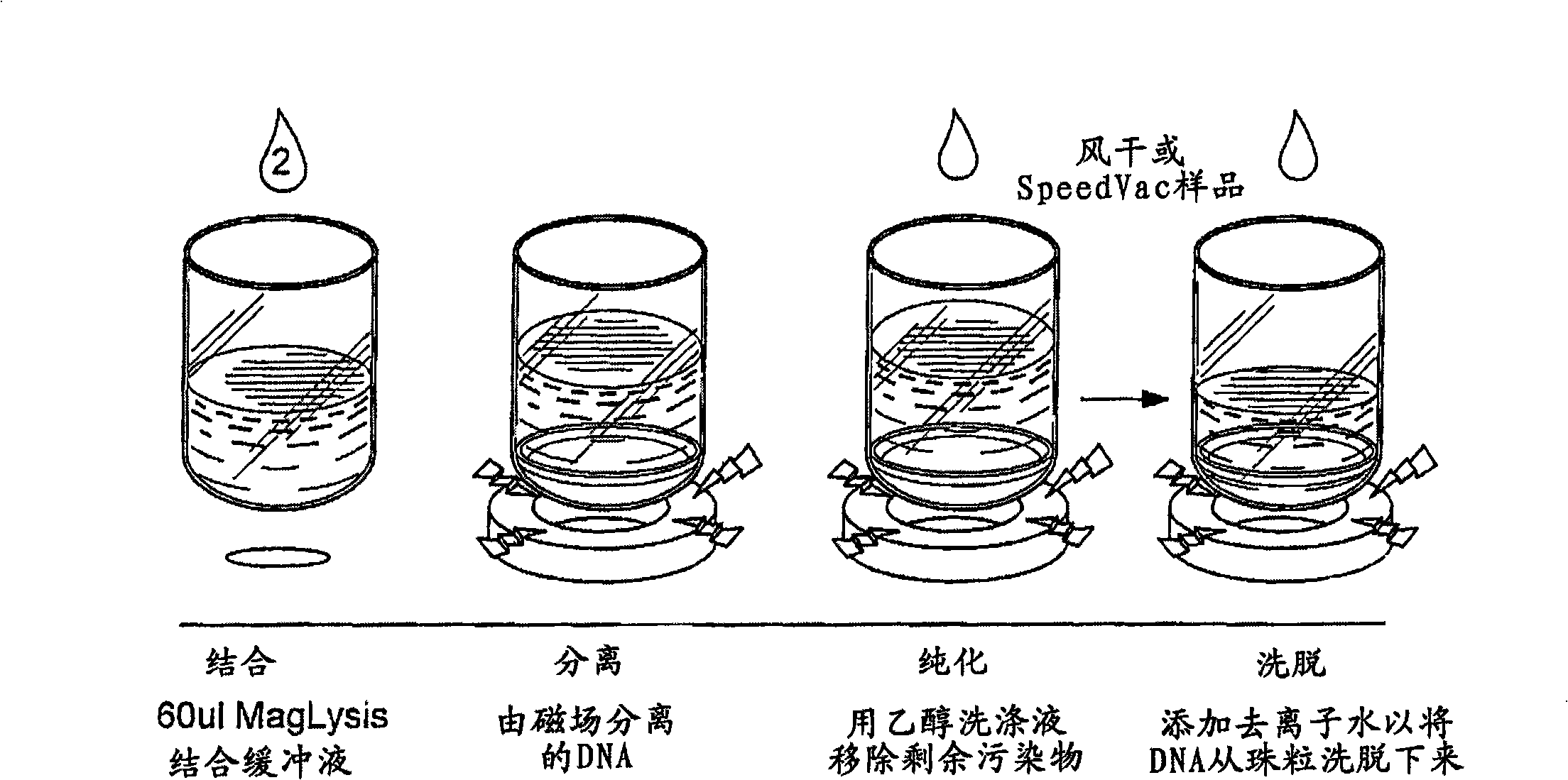
[0078] Example 1 One Embodiment of the Protocol for Isolating Nucleic Acids with Lower Molecular Weight than Genomic Nucleic Acids in Cells
[0079] The following schemes are described in figure 1 middle.
[0080] growth of bacterial cultures
[0081] 1. Pipette 200 L of 2×YT Bacterial Growth Medium containing appropriate antibiotics into each well of a 300 l Costar 96-well round bottom culture plate (Catalog No. 3750).
[0082] 2. The wells were implanted with a single plasmid (DH10B, DH5alpha: Invitrogen) containing E. coli bacterial colonies. Growth cultures can be seeded directly from agar lawns or glycerol stocks.
[0083] 3. Cover the culture plate with a perforated lid and shake vigorously (eg 300 rpm) at 37°C for 16 hours.
[0084] Purification procedure
[0085] 1. Add 60 L of paramagnetic lysis buffer (0.4 N NaOH, 2% SDS, 0.0016% solids Agencourt COOH magnetic particles) solution to each well of the source plate and mix by shaking or tilting.
[0086] 2. Add 60...
Embodiment 2
[0108] Example 2 Separation of Exogenous Nucleic Acids with Lower Molecular Weight than Genomic Nucleic Acids in Bacterial Cells
[0109] Methods and Materials
[0110] Cloning and Purification: Chimpanzee genomic DNA was sheared, end-repaired with T4 polymerase and Klenow (NEB), and cloned in the pOT bacterial vector. DH10B cells (Invitrogen) were electroporated and plated on 25 μg / ml chloramphenicol agar and grown overnight. Colonies were picked into 200 μl 2XYT, 50 μg / ml chloramphenicol broth with a Gentix Qpix and grown for 16 hours. Clones were purified in growth plates on a Beckman FX robotic platform.
[0111] Purification method
[0112] The method described herein (OneStep Prep) was compared to the method described in US Patent No. 6,534,262 (McPrep).
[0113] 24 samples were purified using two different purification methods (McPrep and the single-step purification method described herein). Control samples were loaded on an agarose gel to compare recovery ( fig...
Embodiment 3
[0143] Example 3 Isolation of Nucleic Acids with Lower Molecular Weight than Genomic Nucleic Acids in Horse Whole Blood Cells
[0144] Materials and methods
[0145] source
[0146] Horse whole blood was obtained in a 1:1 ratio with Alsevers anticoagulant (2.05% dextrose, 0.5% sodium citrate, 0.055% citric acid, 0.42% sodium chloride).
[0147] Purification method
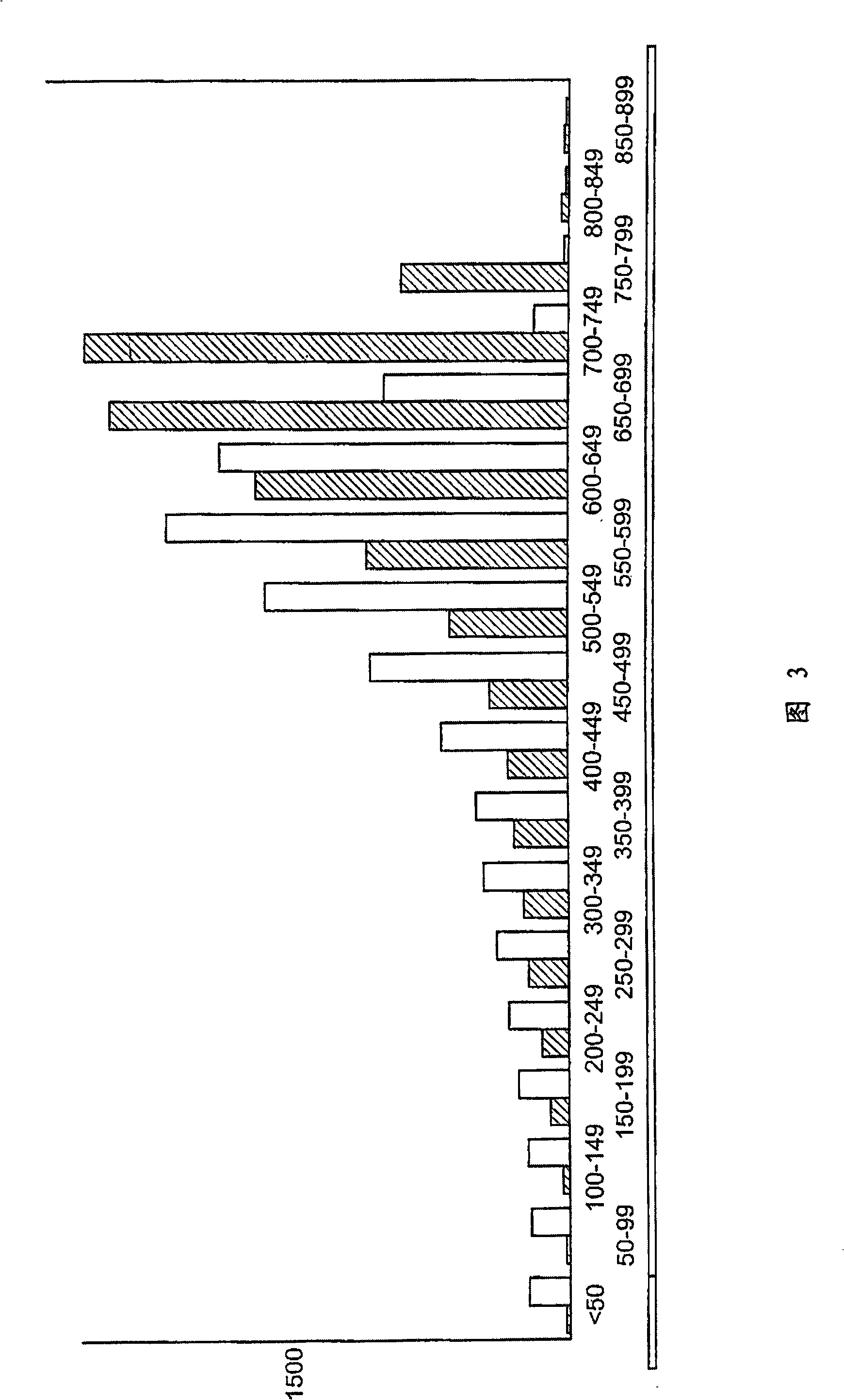
[0148] In addition to 80 μl of 100% isopropanol, 60 μl of paramagnetic lysis buffer (0.4 NaOH, 2% SDS, 160 mg / L Seradyn COOH paramagnetic beads) was added to the cell culture and mixed by tilting 15 times. Samples were separated on Agencourt magnet plates (1000 Gauss) for 15 minutes. The supernatant was removed and the separation beads were washed 5 times with 70% ethanol. using ddH 2 After O (Sigma) elution, samples were run on 96 E-Gel (Invitrogen) to assess relative DNA recovery ( Figure 4 ). Samples were also subjected to Pico Green analysis (Molecular Probes) and the average DNA recovery was 0.5 ng / μl ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Conductivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com