Bicistronic specific DNA utilizing lacZ alpha oligopeptide encoding gene as second gene encoding frame and application of bicistronic specific DNA
A technology of gene coding and DNA molecules, which is applied in the biological field and achieves the effect of great application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Embodiment 1, construction of recombinant plasmid
[0049] 1. Synthesize the double-stranded DNA molecule shown in sequence 1 of the sequence listing.
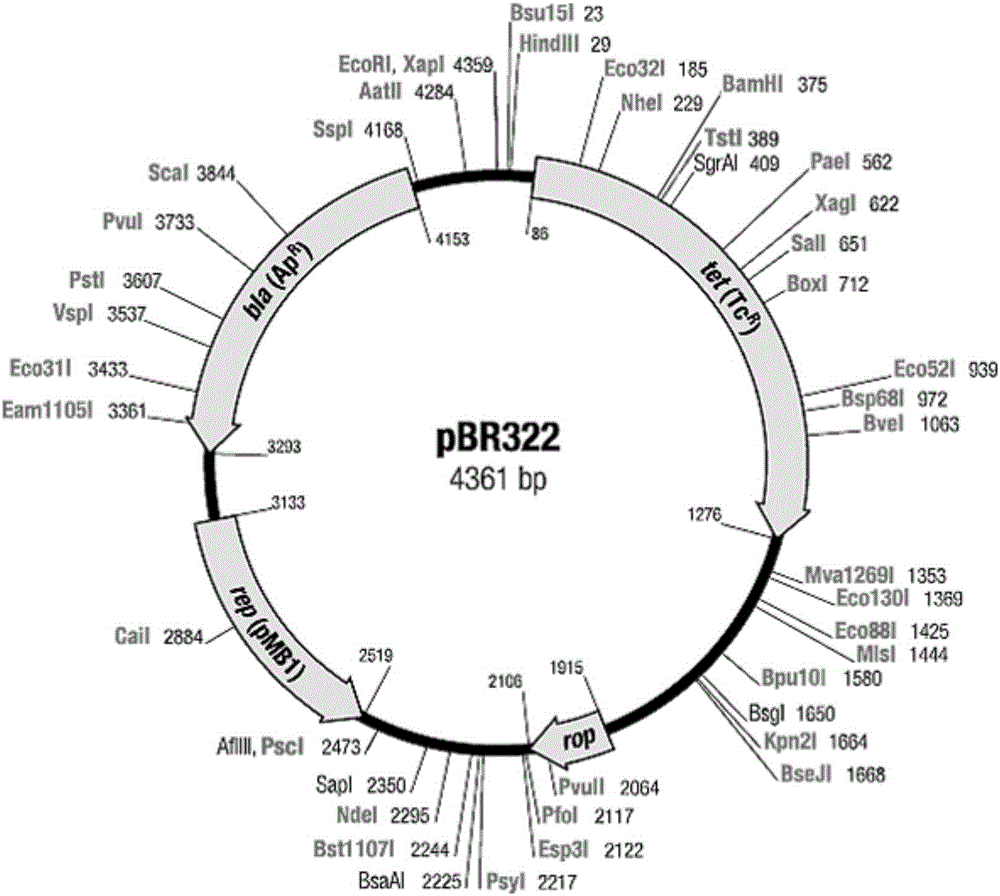
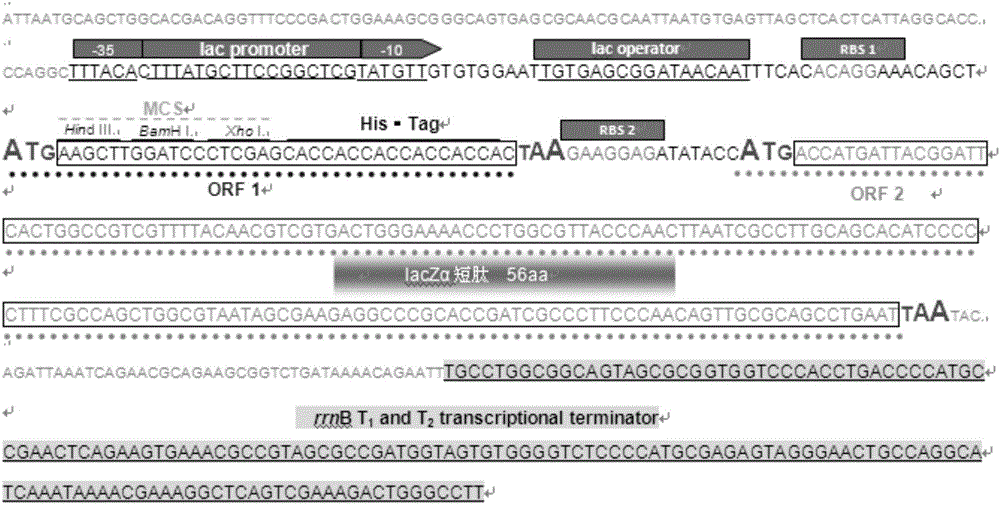
[0050] In sequence 1 of the sequence listing, the 9th-14th nucleotides are the recognition sequence of the restriction endonuclease Nde I, the 112th-141st nucleotides are the lac promoter region, and the 146th-168th nucleotides are lac In the control region, the 173rd-177th nucleotides are RBS1, the 186th-188th nucleotides are the start codon of the gene coding frame Ⅰ, and the 189th-206th nucleotides are the MCS region (the sequence is cut by HindⅢ recognition sequence, BamHI restriction recognition sequence and XhoⅠ restriction recognition sequence), the 207th-224th nucleotide is His 6 The coding sequence of the tag, the 225th-227th nucleotide is the stop codon of the gene coding frame I, the 228th-234th nucleotide is RBS2, and the 242-412th nucleotide is the coding sequence of the lacZα short peptide ( The 242nd-24...
Embodiment 2
[0058] Embodiment 2, construct recombinant bacteria
[0059] The recombinant plasmid pBR-ORF-lacZα was introduced into Escherichia coli Top10 to obtain recombinant bacteria I.
[0060] The recombinant plasmid pBR-ORFmu1-lacZα was introduced into Escherichia coli Top10 to obtain recombinant bacteria II.
[0061] The recombinant plasmid pBR-ORFmu2-lacZα was introduced into Escherichia coli Top10 to obtain recombinant bacteria III.
[0062] The recombinant plasmid pBR-ORFmu3-lacZα was introduced into Escherichia coli Top10 to obtain the recombinant strain IV.
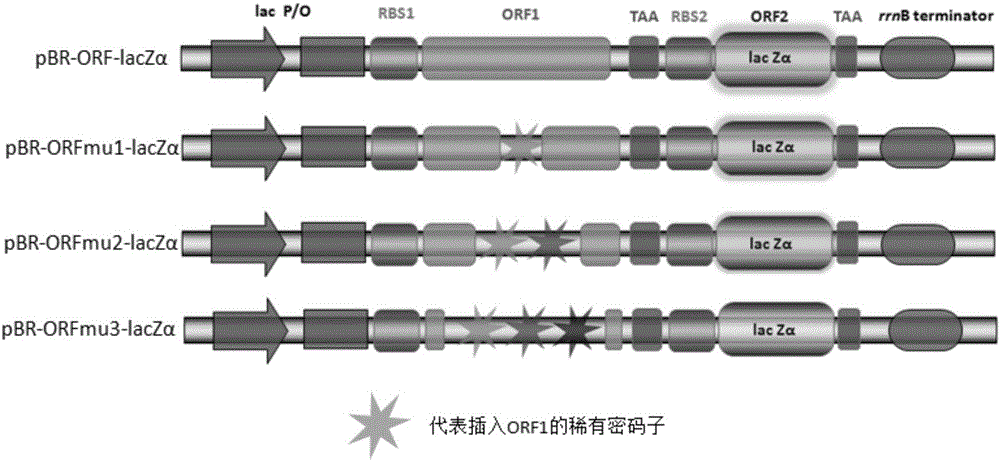
[0063] The schematic diagram of the components of each recombinant plasmid is shown in image 3 .
[0064] Rare codons, especially consecutive rare codons, are resistant to gene expression. When rare codons appear in the coding frame of the first gene, the translation level of the coding frame of the second gene will decrease compared to the situation without rare codons, that is, the expression level of the reporter g...
Embodiment 3
[0065] Example 3, Detection of expression of gene coding frame I in recombinant bacteria by color or OD value
[0066] 1. Test treatment
[0067] The recombinant bacteria I, recombinant bacteria II, recombinant bacteria III and recombinant bacteria IV constructed in Example 2 were respectively used as the bacteria to be tested, and the following operations were performed:
[0068] 1. Inoculate the bacteria to be tested into the liquid LB medium containing 50 μg / mL ampicillin, culture at 37°C with shaking at 250 rpm until OD 600nm Value = 0.6.
[0069] 2. After completing step 1, add IPTG and X-gal to the culture system (the concentration of IPTG in the culture system is 0.5mM, and the concentration of X-gal in the culture system is 0.04g / 100ml), shake at 37°C and 250rpm Incubate for 4 hours.
[0070] 3. After completing step 2, take the entire culture system, centrifuge at 3500rpm, take the supernatant, and take pictures.
[0071] 4. Take the supernatant obtained in step 3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com