Fluorescence in situ hybridization (FISH) image parallel processing and analysis method
A fluorescence in situ hybridization and image processing technology, applied in the field of biomedicine, can solve the problems of inaccurate detection of the border of adherent nuclei, increased image processing running time, and wrong counting of fluorescent marker points.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0145] Example 1 FSIH detection of tumor cells
[0146] Take tumor cells, and perform FISH staining on chromosomes 1 (green), 7 (red), 8 (orange), and 17 (blue) using fluorescent dyes with the following colors, respectively, and then perform FISH imaging.
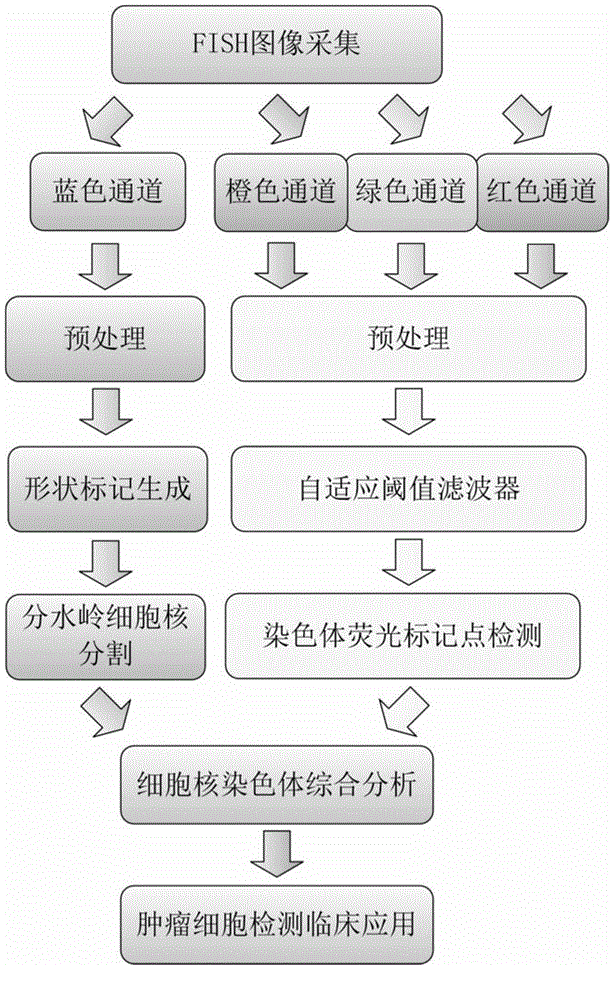
[0147] Using the FISH image parallel processing and analysis method based on the adaptive shape marker watershed algorithm of the present invention, the obtained FISH image is analyzed, such as figure 1 It mainly includes the following steps:
[0148] 1. Preprocess FISH images derived from tumor tissue sections, including image decomposition and grayscale, to obtain grayscale images containing only nuclei and grayscale images containing only orange, green, and red chromosomal fluorescent markers respectively. , the specific implementation process is divided into five steps as follows (see appendix figure 2 ):
[0149] 1.1 Read the original FISH color RGB image in the form of a three-dimensional matrix, where the third d...
Embodiment 2
[0175] Example 2 FSIH detection of circulating tumor cells
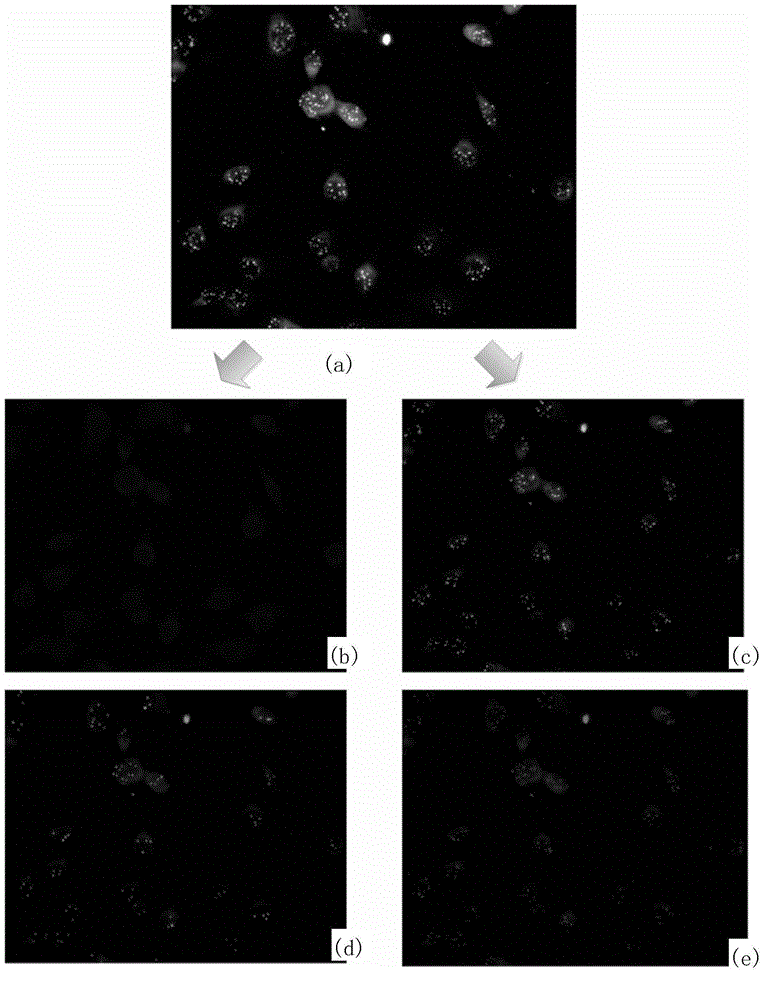
[0176] In Example 2, after whole blood cells were lysed red blood cells, a large number of leukocytes were removed by negative enrichment, nuclei were stained with DAPI, chromosomes were stained with orange-labeled probes, and leukocytes were stained with red-labeled antibodies. Circulating tumor cells were detected using substantially the same method as in Example 1. In Example 2, the circulating tumor cell images containing 60 cells were detected, and the processing speed was 5 seconds per image on a common desktop computer with 2.4GHz CPU and 4GB memory, and the detection accuracy of tumor cells was 92.9%, which was the same as the conventional method. Compared with this, the detection speed and accuracy are significantly improved.
[0177] The schematic diagram of the present invention for FISH detection of circulating tumor cells is as follows Figure 8 where (a) is the blue monochromatic image including the n...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com