Quick experimental animal resource quality monitoring method
A technology of experimental animals and resources, applied in the field of biology, can solve the problems of low specificity and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Embodiment 1, formulation of experimental technical route and utilization of positive standard bacterial strains to screen primers
[0060] 1. Establishment of PCR method for positive control
[0061] The target microorganisms to be detected in this experiment are: Staphylococcus aureus, beta-hemolytic streptococcus, and Klebsiella pneumoniae.
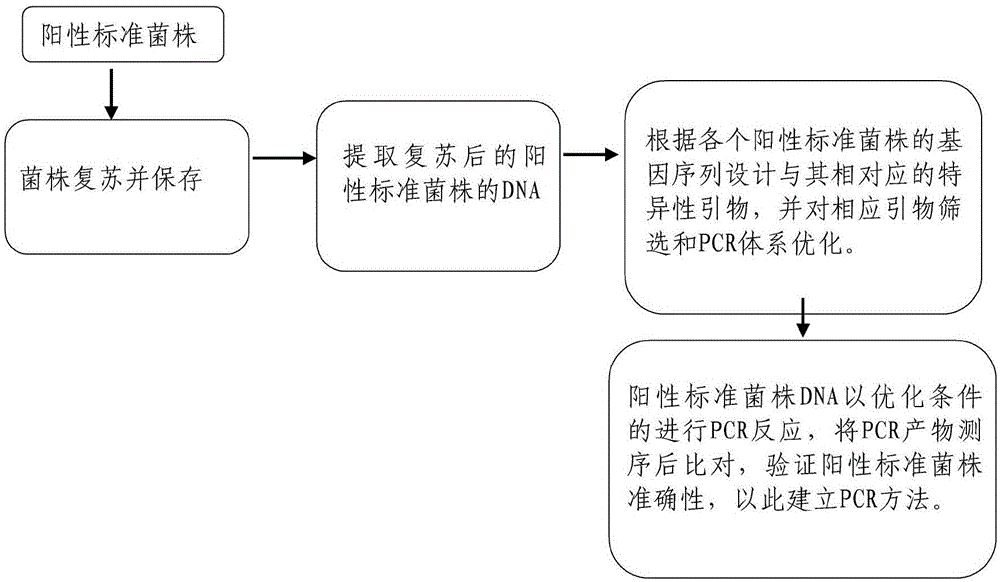
[0062] Purchase the above three standard strains from the China Medical Microbial Culture Collection Center (CMCC) as positive controls, resuscitate these three standard strains, extract the DNA of the revived standard strains, and design corresponding to each standard strain according to the gene sequence The specific primers were selected, the corresponding primers were screened and the PCR system was optimized, and the PCR products were sequenced and compared to verify the accuracy of the positive standard strains, so as to establish the PCR method. Process such as figure 1 .
[0063] 2. Primer screening
[0064] After a ...
Embodiment 2
[0084] Example 2, positive standard strain PCR result product sequencing result BLAST
[0085] Electrophoresis and sequencing were performed on the obtained positive standard strain PCR products. The obtained sequencing results were subjected to BLAST. The parameters are as follows:
[0086] Score is the alignment score, the higher the similarity between two sequences;
[0087] The smaller the E Value, the more credible it is;
[0088] Identities is consistency, which means that the two sequences maintain the same ratio.
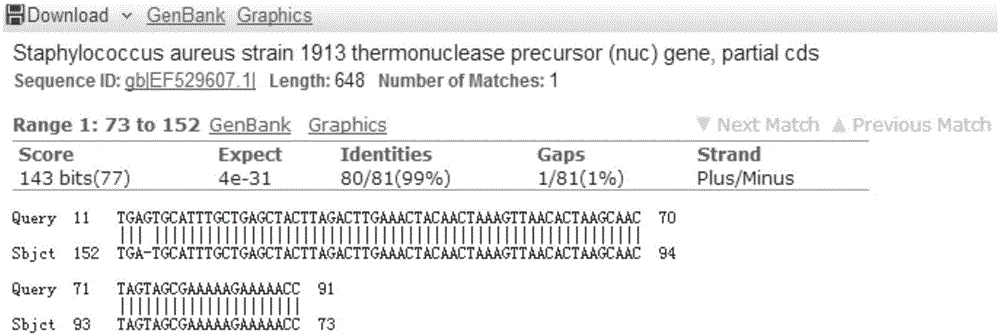
[0089] 1. Staphylococcus aureus primer 1
[0090] Using Staphylococcus aureus primer 1, the target band 122bp was obtained. Blast results such as image 3 shown.
[0091] 2. Staphylococcus aureus primer 2
[0092] Using Staphylococcus aureus primer 2, the target band 132bp was obtained. Blast results such as Figure 4 shown.
[0093] 3. Klebsiella pneumoniae primer 1
[0094] Klebsiella pneumoniae primer 1 was used to obtain the target band 368bp...
Embodiment 3
[0102] Example 3, establishment of a rapid detection method for frozen resources
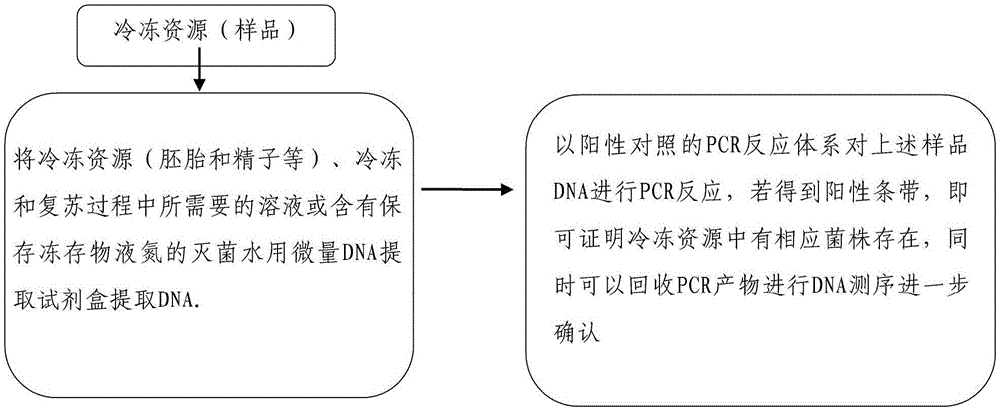
[0103] The frozen resources (embryo and sperm, etc.), the solution required in the freezing and recovery process, and the sterilized water containing liquid nitrogen for preserving frozen storage were used to extract DNA with a micro DNA extraction kit (purchased from QIAGEN).
[0104] PCR primers and corresponding PCR conditions and PCR reaction system for Staphylococcus aureus, Klebsiella pneumoniae, beta-hemolytic streptococcus provided in the foregoing embodiment 1 are carried out to the above-mentioned sample DNA for PCR reaction, if a positive strip is obtained If there is a band, it can prove that the corresponding strain exists in the frozen resources, and at the same time, the PCR product can be recovered for further confirmation by DNA sequencing.
[0105] Operation process such as figure 2 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com