Biological preparation method of (S)-1-(2,6-dichloro-3-fluorophenyl)ethanol
A technology of fluorophenyl and fluoroacetophenone, which is applied in the field of biopharmaceuticals, can solve the problems of unfavorable extraction efficiency and operation process, complicated reaction system and high production cost, and achieves convenient industrial production, simple operation process and high ee value. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Construction of co-expression recombinant bacteria
[0028] The fully synthetic KRED182 gene was used as a template for PCR amplification, and restriction sites were introduced at both ends (forward primer was introduced into NcoI restriction site, reverse primer was introduced into HindIII restriction site), and NcoI and HindIII were used for its enzymes. cut and recovered the KRED gene fragment. At the same time, the plasmid of the pRSF-Duet vector was extracted and digested with NcoI and HindIII, and the digested vector fragment was recovered. The KRED gene fragment and the vector fragment were ligated using T4 ligase, transformed into E. coli BL21 (DE3), coated on a Kan-resistant plate, and cultured in a 37°C incubator. After the transformants were grown, several single clones were picked for colony PCR verification, and the positive single clone was selected for subsequent experiments, and the strain was named KRED-1 bacteria. Then, the KRED bacterial p...
Embodiment 2
[0029] Example 2 Preparation of free cells and immobilized whole cells of co-expressed recombinant bacteria
[0030] Co-expressed free cells can be obtained by fermentation and culture. The fermentation medium uses 2YT medium, which contains 16g / L peptone, 10g / L yeast powder, 5g / L NaCl, and 2g / L glycerol. The specific steps are as follows: firstly inoculate the strain into a small shaker flask (250ml capacity) containing 50mL of culture medium, cultivate the strain at 37°C for 12h on a shaker to activate the strain, and then inoculate the activated bacteria into a small shaker with a capacity of 250ml. In a 400mL medium shake flask (capacity 1L, with baffle), the inoculum volume is 8ml, incubate at 37°C on a shaker until the OD600nm reaches 0.6-1.0, add IPTG to a final concentration of 0.1mM, and shake at 25°C for induction After 20 h of expression, the co-expressing cells were collected by centrifugation, which were called free cells.
[0031] Weigh 300g of polyvinyl alcohol...
Embodiment 3
[0032] Example 3 Preparation of (S)-1-(2,6-dichloro-3-fluorophenyl)ethanol by transformation of co-expressed free cells
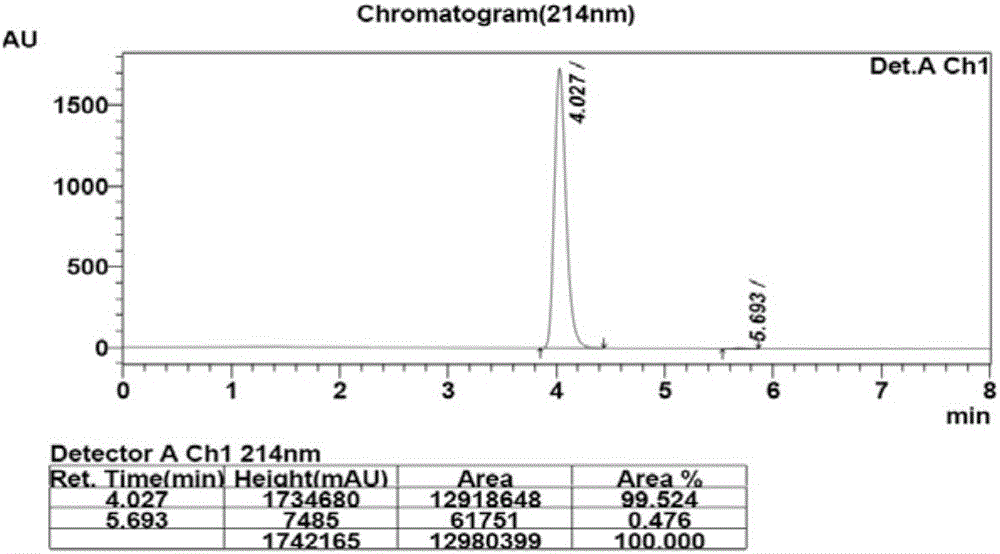
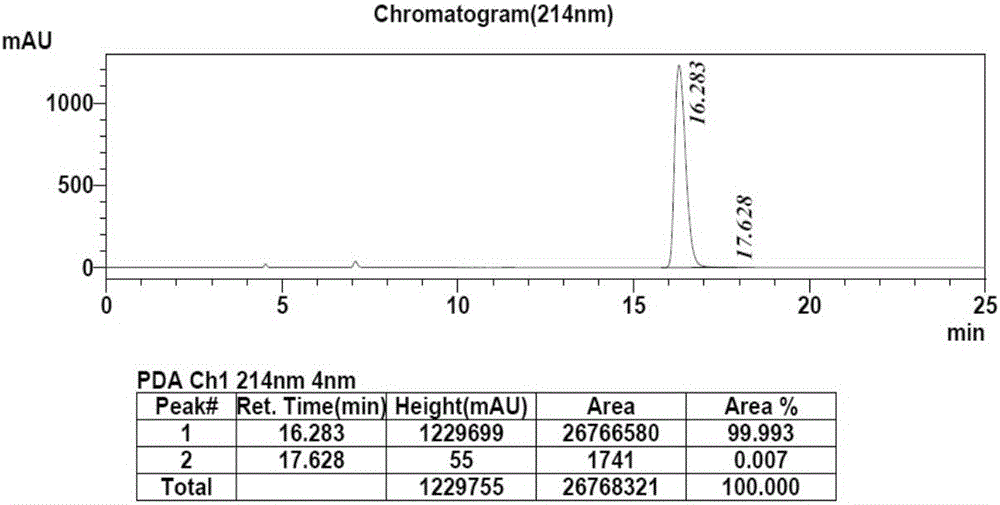
[0033] 100mM phosphate buffer (10mL, pH=6.0), glucose (3.0g) and substrate 2,6-dichloro-3-fluoroacetophenone (3.0g) were added to a 25mL reaction vessel, and the volume was adjusted to 15mL , co-expressed free cells (750mg) after stirring evenly, magnetic stirring reaction at 35 ℃, Na 2 CO 3(20%, w / v) pH of the reaction was controlled at about 6.0, and the reaction progress was detected by TLC. After the reaction, the cells were removed by centrifugation, extracted three times with equal volumes of ethyl acetate, the organic phases were combined, dried over anhydrous sodium sulfate, and spin-dried under reduced pressure to obtain the product (S)-1-(2,6-dichloro-3-fluorobenzene) base) ethanol 2.84g, the molar yield of the product is 94%, and the ee value is greater than 99.9%. HPLC detects the conversion rate and the ee value of the product, the conversio...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| enantiomeric excess | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com