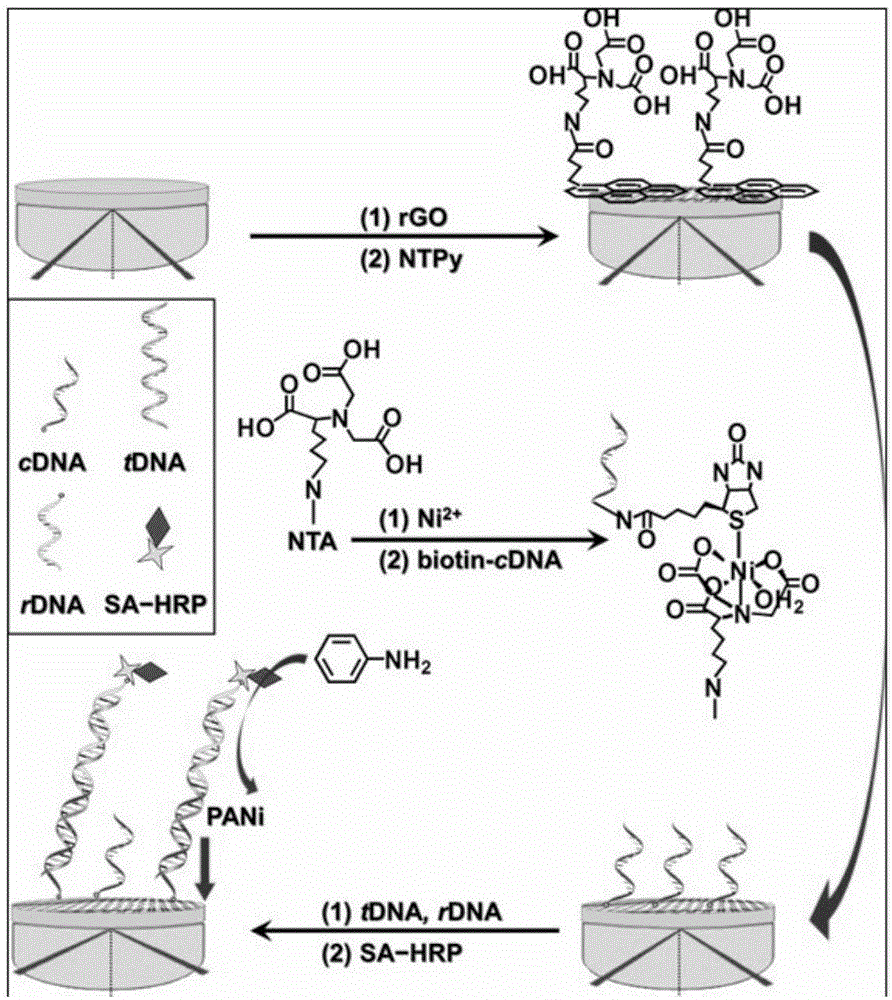
Nucleic acid detection method based on surface plasma resonance technology
A technology of surface plasma and detection method, applied in the field of nucleic acid analysis, can solve the problems of insufficient detection sensitivity, low probe density, poor binding stability, etc., and achieve the effects of simple operation, low cost and stable assembly structure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Electrodeposition of reduced graphene oxide nanosheets on the surface of the SPR chip, the specific process is as follows:
[0048] (1) The bare SPR chip was prepared in piranha acid (concentrated H 2 SO 4 : Concentrated H 2 o 2 =1:3) after soaking for 10min, then in boiling H 2 o 2 Mixed solution with ammonia water (H 2 o 2 The mass concentration of ammonia and ammonia water is 5%) and soaked for 10min, and finally rinsed twice with ultrapure water and ethanol. 2 Blow dry for later use;
[0049] (2) Add 0.5mg·mL -1 GO solution in 0.1M Na 2 SO 4 Ultrasonic for 2h, get 0.5mg·mL -1 GO dispersion;
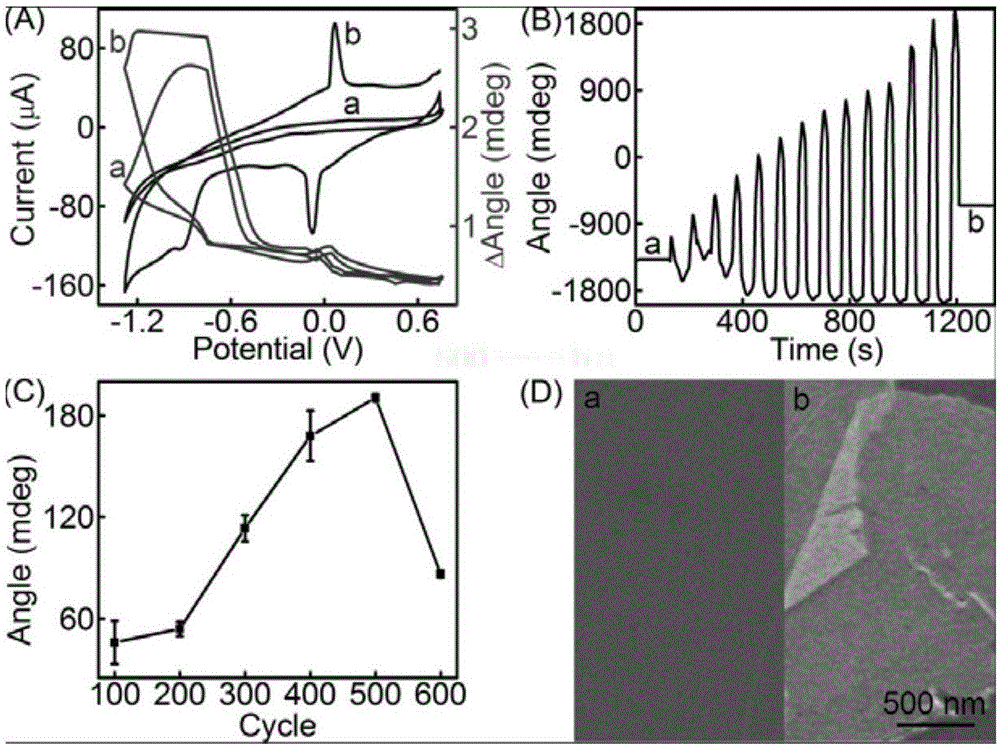
[0050] (3) Cyclic voltammetry scanning on-line electrical reduction of GO, the cycle voltage is -1.5 ~ +0.5V, and the cycle speed is 50mVs -1 , for 1 to 600 cycles, to obtain an SPR chip with electrodeposited reduced graphene oxide nanosheets on the surface.
[0051] figure 2 A is the cyclic voltammetry curve, where (a) is the 1st time, (b) is the 500th time, an...
Embodiment 2
[0053] The underlying construction of cDNA / NTPy / rGO and its hybridization performance, the synthesis steps are:
[0054] (1) Micropipette 0.2 mL of dimethylformamide containing 2 mM aminotriacetic acid (NTA)-grafted perylene derivative (NTPy) into the container and spread it on the electrodeposited surface after 500 cycles. On the SPR chip of graphite oxide, and incubate for 1h to lay the foundation of the π-stacking scaffold, and repeatedly pump in / out DMF to wash away the weak adsorbate;
[0055] (2) Continuously, 0.5mL, 10mM Ni(ClO 4 ) 2 The acetic acid buffer solution was drop-coated on the above-mentioned treated chip, and left to stand for 2-6 hours to complex Ni 2+ and rinse with acetic acid buffer;
[0056] (3) The divisible mixed eluent of 1 μM cDNA and 5 μM ethanolamine blocking agent (blocking non-specific binding sites) is introduced onto the chip obtained in step 2, and the construction of the bottom layer is completed through layer-by-layer assembly to form cD...
Embodiment 3
[0060] The non-covalent assembly of dsDNA / NTPy / rGO, the reaction process is as follows:
[0061] Inject 100 μL, 1 pM tDNA into the detection cuvette, and perform initial biometric recognition with the cDNA / NTPy / rGO chip synthesized in Example 2, to realize binding and capture of target DNA, form dsDNA / NTPy / rGO, and complete the detection step.
[0062] Figure 5 Atomic force microscopy (AFM) images of rGO / Au (A), NTPy / rGO / Au (B) and dsDNA / NTPy / rGO / Au (C). Figure 5 A shows that within the scanned area of 2 × 2 μm, rGO maintains a broken morphology, which is consistent with figure 2 D is equivalent, rGO fragments occupy the entire area, and have a broad size distribution, with an average height of about 2nm in the plane (Z-offset=0nm). Figure 5 B shows that NTPy is in situ adsorbed on rGO, given the van der Waals radius of NTA, the average height of the sampling interval is increased by 6 nm, indicating that rGO and NTPy are tightly connected through π conjugation. Because...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com