Specific molecular markers for detection of Aegilops comosa 2M, 3M, 6M and 7M chromosomes in wheat, kit and method
A molecular marker and chromosome technology, applied in the field of molecular biology, can solve the problem of lack of awn goat grass and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
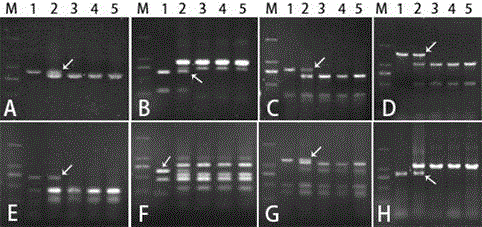
[0052] Designing and screening PLUG primers for homology groups 2, 3, 6, and 7 of chromosomes 41 and 2, 3, 6, and 7 using A. chinensis, Chinese spring-A. For pairs, 84 pairs, 53 pairs and 73 pairs, the amplified products were digested with restriction endonuclease HaeIII and TaqI respectively, and the PCR amplification products from the same primer (undigested, HaeIII digested and TaqI digested three case) agarose gel electrophoresis, it was found that the CD913720 of the second homologous group of chromosomes ( figure 1 A) and CK171581 ( figure 1 B), CD491237 of the third homology group of chromosome ( figure 1 C) and CK207576 ( figure 1 D), CK168221 of the sixth homologous group of chromosome ( figure 1 E) and CK206466 ( figure 1 F), CK214770 of the seventh homologous group of chromosome ( figure 1 G) and CK203272 ( figure 1 H) Additional polymorphic bands can be amplified in A. chinensis and Chinese spring-A. 650bp, 1200bp and 770bp ( figure 1 Arrows in A-H), howeve...
Embodiment 2
[0056] Up to now, since there is no report on the creation of the 1M addition line of the Chinese spring-A. g Chromosome donors may have undergone partial recombination or variation of chromosomes during the polyploidization and evolution of the species, resulting in slightly different chromosomes from A. chinensis and the M chromosome of A. , with M g Indicated to show the difference. Therefore, in the absence of the Chinese spring-A. g The additional system is used for marker positioning.
[0057] In order to locate the above polymorphic bands from A. chinensis on one or several chromosomes of A. chinensis 1M-7M, the 8 pairs of PLUG primers screened above were used to pair Chinese spring-A. Grass Diploid, Chinese Spring, Chinese Spring - Goat Grass 1M g Addition line, Chinese Spring-Dingmang goat grass 2M addition line, Chinese spring-Dingmang goat grass 3M addition line, Chinese spring-Dingmang goat grass 4M addition line, Chinese spring-Dingmang goat grass 5M addition ...
Embodiment 3
[0063] In order to verify the practicability of the above 8 markers, the 8 pairs of primers screened above were used to amplify the 4 populations respectively to verify the practicability of the markers.
[0064] Among them, CD913720 and CK171581 of the second chromosome homology group were used to amplify P1-P42 progeny materials of the Chinese Spring-Dingmis chinensis 2M addition line / Chinese Spring hybrid F2 progeny, and the results showed that both CD913720 and CK171581 could be used in wheat- Amplified polymorphisms in the amphidiploid, wheat-A. acanthus 2M addition line, P2-P6, P16-P20, P25, P26, P28, P31, P32, P36, P39, P40 and P42 Marked CD913720-PLUG and CK171581-PLUG, but no corresponding polymorphic bands were amplified in Chinese Chunhe P1, P7-P15, P21-P24, P27, P29, P30, P33-P35, P37, P38 and P41 (Table 2). Among them, the amplification results of CD913720 on P1-P21 are as follows image 3 As shown in A, the amplification result of CK171581 on P22-P42 is as foll...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com