Method for detecting number of arbuscular mycorrhizal fungi in wheat rhizosphere soil based on real-time fluorescence quantification PCR
An arbuscular mycorrhizal fungi, real-time fluorescence quantitative technology, applied in the biological field, can solve the problems of arbuscular mycorrhizal fungi counting and other problems that have not yet been seen in qPCR technology, and achieve a simple and accurate identification method, high amplification efficiency, and high linearity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] (1) Construct the target plasmid
[0039] According to the 28s rDNA conservative sequence of ribosomal DNA of common fungi in the arbuscular mycorrhizal family and soil on the NCBI website, the BioEdit software was used for comparison and then the upstream and downstream primers for specifically amplifying arbuscular mycorrhizal specific fragments were designed such as SEQ ID No. 1 and SEQ ID No.2.
[0040] The applicant took samples from the Liuhe Transgenic Experiment Base of Jiangsu Academy of Agricultural Sciences to obtain transgenic wheat and its receptor rhizosphere soil, using UltraClean TM soil DNA Isolation Kit, extract total DNA from soil and dissolve total DNA in ddH 2 In O; use SEQ ID No.1 and SEQ ID No.2 as the upstream and downstream primers, and the total soil DNA as the template to amplify the target band:
[0041] The PCR reaction system is 25 μL, in which rTaq enzyme 0.125 μL, 10×PCR buffer 2.5 μL, dNTP (dATP, dTTP, dCTP, dGTP each 2.5mM) 2 μL, MgCl 2 (25...
Embodiment 2
[0058] Example 2 Sensitivity detection of fluorescence quantitative PCR detection system
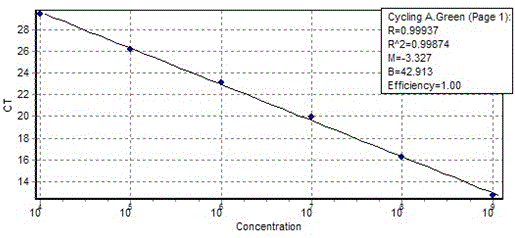
[0059] For the standard plasmid (2.0×10 10 copies / µl) 10 times dilution, 1*10 8 copies / µL-1*10 3 copies / µL, use SEQ ID No.1 and SEQ ID No.2 for fluorescence quantitative amplification to detect the sensitivity of amplification. Refer to Example 1 for specific reaction system and reaction conditions.
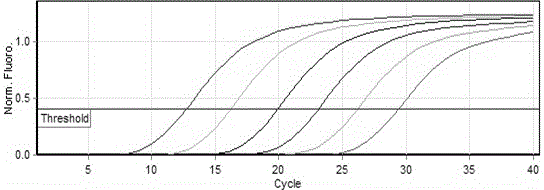
[0060] Get the amplification curve as Figure 4 As shown, Figure 4 Among them, 1, 2, 3, 4, 5, 6, 7, 8 represent the standard plasmid stock solution in turn, 10 -1 ,10 -2 ,10 -3 ,10 -4 ,10 -5 ,10 -6 ,10 -7 Double dilution; perform agarose gel electrophoresis on the amplification results, such as Figure 5 As shown, Figure 5 Among them, M: DL 2000 maker, 1, 2, 3, 4, 5, 6, 7, 8 represent the standard plasmid stock solution, 10 -1 ,10 -2 ,10 -3 ,10 -4 ,10 -5 ,10 -6 ,10 -7 Fold dilution; by Figure 4 It can be seen that in the 10 -6 Before the double dilution, the amplification curve still showed an...
Embodiment 3
[0061] Example 3 Analysis of growth dynamics of arbuscular mycorrhizal fungi in rhizosphere soil during the main growth period of transgenic wheat
[0062] The genetically modified wheat and its recipients are planted in the Liuhe Transgenic Experimental Base of Jiangsu Academy of Agricultural Sciences. Each variety (line) has 4 replicates, and a random block design. The area of each plot is 10 (length) × 6 (width) m 2 .
[0063] The wheat rhizosphere soil was collected by the diagonal five-point sampling method at the same place during the sowing, jointing, filling, and maturation periods of wheat growth, and DNA was extracted using the specific primers and amplification system provided in Example 1. Amplify and calculate according to the standard curve to obtain the bacterial content of the initial arbuscular mycorrhizal fungi in the soil sample.
[0064] The established Real-time qPCR was used to detect the number of arbuscular mycorrhizal fungi in the rhizosphere soil during th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com