Molecular cloning method based on CRISPR/Cas9 and homologous recombination of saccharomyces cerevisiae cell endogenous genes
A technology of Saccharomyces cerevisiae cells and molecular cloning, applied in the field of molecular cloning based on CRISPR/Cas9 and endogenous homologous recombination of Saccharomyces cerevisiae cells, which can solve problems that have not been proposed, that have not considered the efficiency of multi-fragment splicing, and have not been mentioned.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
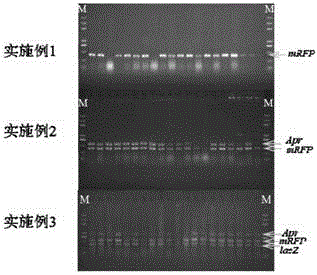
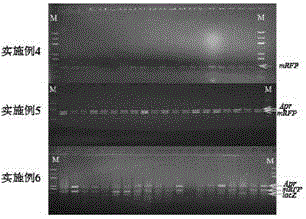
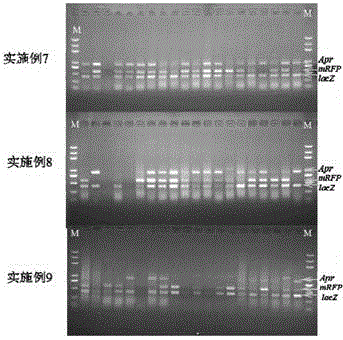
Embodiment 1-3
[0166] Embodiment 1-3: Insert 1-3 target fragments into different positions of the initial vector respectively, and replace a replaced fragment at another position of the initial vector with another target segment at the same time; wherein, the homology arm sequence is located adjacent to be connected flank of one of the fragments
Embodiment 1
[0167] Example 1: Insert a target fragment into the initial vector, and replace the replaced fragment at another position of the initial vector with another target fragment
[0168] The purpose of this example is to insert the target fragment mRFP (SEQ.ID.NO: 4) into site IV of the initial vector, and at the same time, insert the fragment ura3 (SEQ.ID.NO: 6 ) was replaced by the target fragment trp1 (SEQ.ID.NO:2).
[0169] (1) Positioning: designed for image 3 The sgDNA sequences of sites I, II and IV (sgDNA-1, sgDNA-2, sgDNA-3, as shown in Table 1). Each sgDNA was ligated into the sgRNA in vitro transcription template (SEQ.ID.NO: 7) by overlapping PCR, and the primers used in overlapping PCR were shown in Table 1.
[0170] The PCR purification kit (Omega-D6492) was used to recover and purify the obtained sgRNA in vitro transcription template.
[0171] The Epicenter ASF3257 commercial kit was used to perform in vitro transcription using T7 RNA polymerase to obtain sgRNAs (...
Embodiment 2
[0185] Example 2: Insert two target fragments into different positions of the initial vector, and replace the replaced fragment at another position of the initial vector with another target fragment
[0186] The purpose of this example is to insert the target fragments mRFP (SEQ.ID.NO: 4) and lacZ (SEQ.ID.NO: 3) into site IV and site III of the initial vector respectively, and at the same time, insert the site The fragment ura3 between I and II was replaced by the target fragment trp1.
[0187] (1) Positioning: by a method similar to that described in Example 1, design and synthesize targeting image 3 The sgRNA sequences of site I, site II, site IV and site III (sgRNA-1, sgRNA-2, sgRNA-3 and sgRNA-4, as shown in Table 1). All obtained sgRNAs were mixed at equal concentrations.
[0188] (2) Digestion: By a method similar to that described in Example 1, use Cas9 nuclease and the sgRNA obtained in step (1) to digest the initial vector in vitro, so that site I, site II, site D...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com