Double T-DNA vector capable of achieving agrobacterium co-transformation and establishment method and application thereof
A technology of co-transformation and Agrobacterium, applied in the field of genetic engineering, can solve the problems of inability to obtain transgenic positive plants, wrong retention, and difficulty in achieving separation, and achieve the effect of simple and rapid elimination.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Construction of Vector A Containing Two Independent Left Border LB Sequences
[0045] 1. Design primers to amplify the terminator NOS and the left border LB
[0046] According to the sequence of the published pCMBIA1302 vector (GenBank: AF234298.1), the primers for the amplification terminator NOS were designed, including restriction enzyme sites BsteII and BamHI, and the sequences were shown in SEQ ID NO: 1, SEQ ID NO: 2; The primers of the left border LB are added, and the primers include the restriction enzyme cutting sites BglII and SphI, wherein BamHI and BglII are homologous enzymes. See SEQ ID NO:3, SEQ ID NO:4 for the sequence. Primers were synthesized by Shanghai Yingjun Company.
[0047] Using pCAMBIA1302 as a template, using SEQIDNO:1 and SEQIDNO:2 as primers to carry out PCR amplification of NOS, i.e. SEQIDNO:5, the target fragment is about 273bp ( figure 1 ). Use SEQIDNO:3 and SEQIDNO:4 as primers to carry out PCR amplification LB, i.e. SEQIDN...
Embodiment 2
[0050] Embodiment 2 contains the construction of two vectors B facing away from the UBI promoter sequence
[0051] 1. Design primers to amplify two independent UBI promoter fragments
[0052] According to the sequence of the published BinaryvectorpGA1611 carrier (GenBank: AY373338.1), design the primers for amplifying two independent UBI promoter fragments, the primers are designed according to the requirements of homologous recombination primers, the sequences are shown in SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9, SEQ ID NO: 10; a restriction site SalI was introduced into primers SEQ ID NO: 8 and 9.
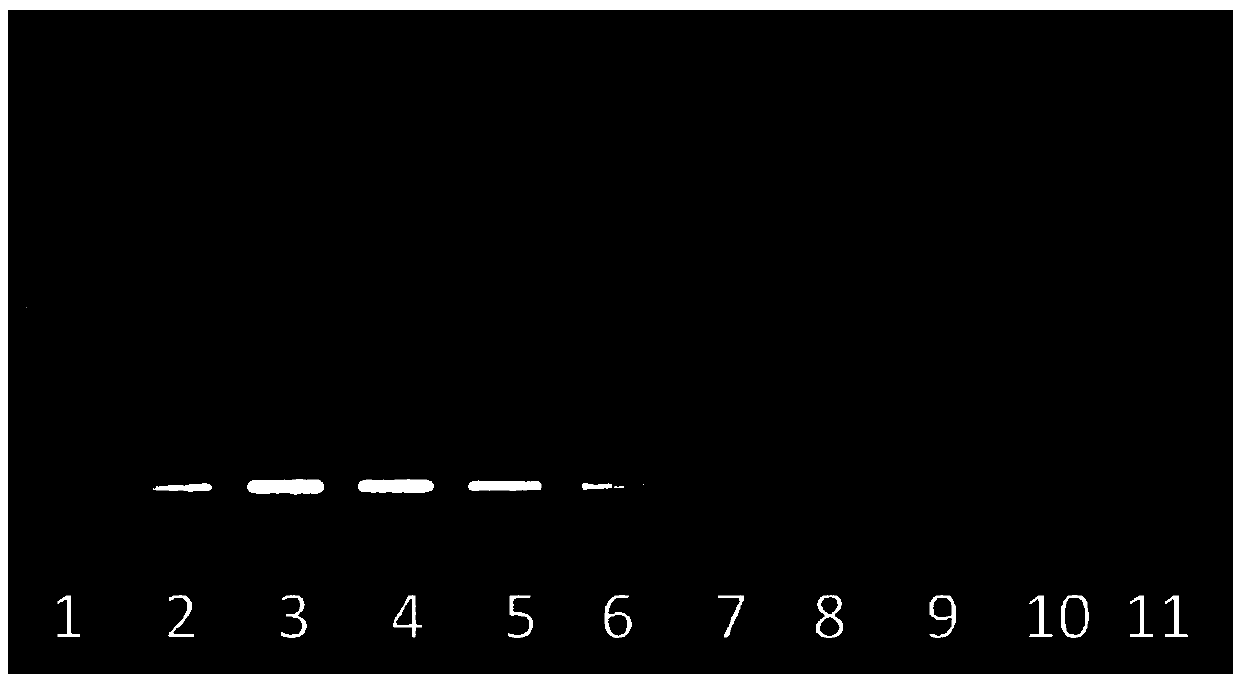
[0053] Using the vector containing the UBI promoter as a template, using SEQIDNO: 7 and SEQIDNO: 8 as primers to perform PCR amplification of UBI-1, i.e. SEQIDNO: 11, the target fragment is about 2050bp ( image 3 ). Use SEQIDNO:9 and SEQIDNO:10 as primers to carry out PCR amplification UBI-2, i.e. SEQIDNO:12, the target fragment is about 2063bp ( image 3 ). The amplified p...
Embodiment 3
[0056] Example 3 Construction of Vector C Containing Two Right Border RB Sequences
[0057] 1. PCR amplification of two independent RB fragments
[0058] According to the sequence (GenBank: AF234298.1) of published pCMBIA1302 carrier, design homologous recombination primer amplification two independent RB-1, RB-2 fragment, primer sequence is respectively SEQIDNO:13, SEQIDNO:14 and SEQIDNO: 15. SEQ ID NO: 16.
[0059] Using pCAMBIA1302 as a template, using SEQIDNO:13 and SEQIDNO:14 as primers to carry out PCR amplification to obtain the RB-1 fragment, the sequence number is SEQIDNO:17, and the target fragment is about 179bp ( Figure 4 ). Carry out PCR amplification with primer and SEQIDNO:15, SEQIDNO:16 to obtain the RB-2 fragment, i.e. SEQIDNO:18, the target fragment is about 173bp ( Figure 4). Gel recovery of the amplified product.
[0060] 2. Homologous recombination ligation and sequencing verification
[0061] The carrier obtained in Example 2 was carried out to Sa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com