Microlunatus phosphovorus engineering strain capable of efficiently accumulating polyphosphate and application thereof
A technology of bacteria and bacteria strains, which is applied in the field of Phosphate-accumulating Lactella engineering bacteria that efficiently accumulates polyphosphate, to achieve the effects of high efficiency, improved performance, and reduced decomposition speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment 1: Continuous error-prone PCR obtains the ppk2 promoter containing mutant bases
[0034] The promoter of the ppk2 gene was mutated in vitro using serial error-prone PCR. Specifically:
[0035] 1. The primers for error-prone PCR are:
[0036] Sequence of upstream primer ppk2ProForward: GTCGAAATGTCGGAAGCCGTGC (SEQ ID NO.2);
[0037] The downstream primer ppk2ProReverse sequence: TAACCCATTGTTCGCCCGGCAG (SEQ ID NO.3).
[0038] (1) PCR reaction system: template DNA 2ul, PCR buffer 5ul, TaqDNA polymerase 1ul, dATP1~3ul, dTTP1~3ul, dGTP1~3ul, dCTP1~3ul, upstream primer 2ul, downstream primer 2ul, 25mMMgCl 2 1~8ul, 5mMMnCl 2 0~1ul, add ultrapure water to 50ul.
[0039] (2) PCR conditions: pre-denaturation at 95°C for 5 minutes, denaturation at 95°C for 30 seconds, annealing at 52-60°C for 30 seconds, extension at 72°C for 2 minutes, 30 cycles, and final extension for 8 minutes.
[0040]The template DNA of the first PCR was derived from S. phosphorus accumulativ...
Embodiment 2
[0042] Embodiment 2: Mutant library construction of Phosphorus acuminosa ppk2 gene promoter
[0043] The shuttle vector pMV306K carrying the upstream and downstream homology arms of the ppk2 gene was constructed, the continuous error-prone PCR products in Example 1 were separated by agarose gel electrophoresis, and the PCR products were recovered, and the above-mentioned shuttle vector pMV306K was recombined in vitro under the action of GibsonCloningMasterMix, Escherichia coli DH5a was transformed, and transformants were picked to extract plasmids for double enzyme digestion identification (pMV306K is a conventional plasmid in the prior art).
[0044] Prepare liquid medium according to the following formula: glucose 0.5g, peptone 0.5g, yeast powder 0.5g, sodium glutamate 0.5g, ammonium sulfate 0.1g, dipotassium hydrogen phosphate 0.44g, magnesium sulfate 0.1g, purified water 1L, pH7 .0, 0.15MPa sterilization for 30min. Each 250ml Erlenmeyer flask is filled with 50ml liquid me...
Embodiment 3
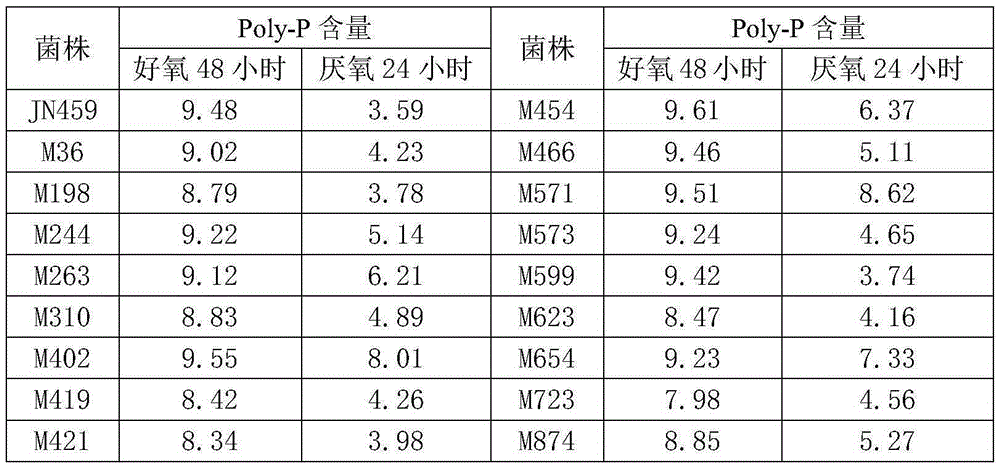
[0049] Example 3: Phosphorus accumulation activity screening of mutant strains
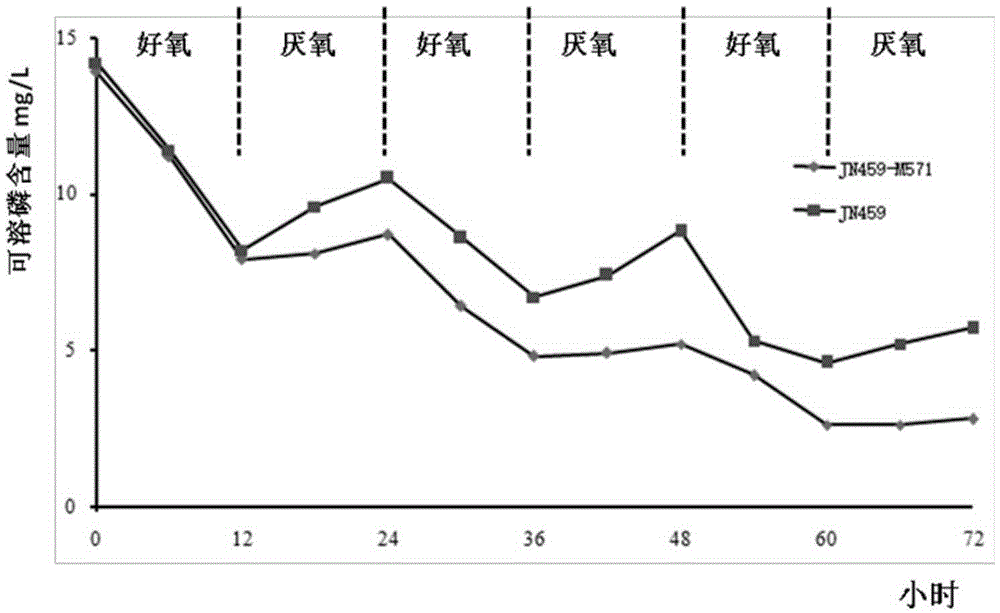
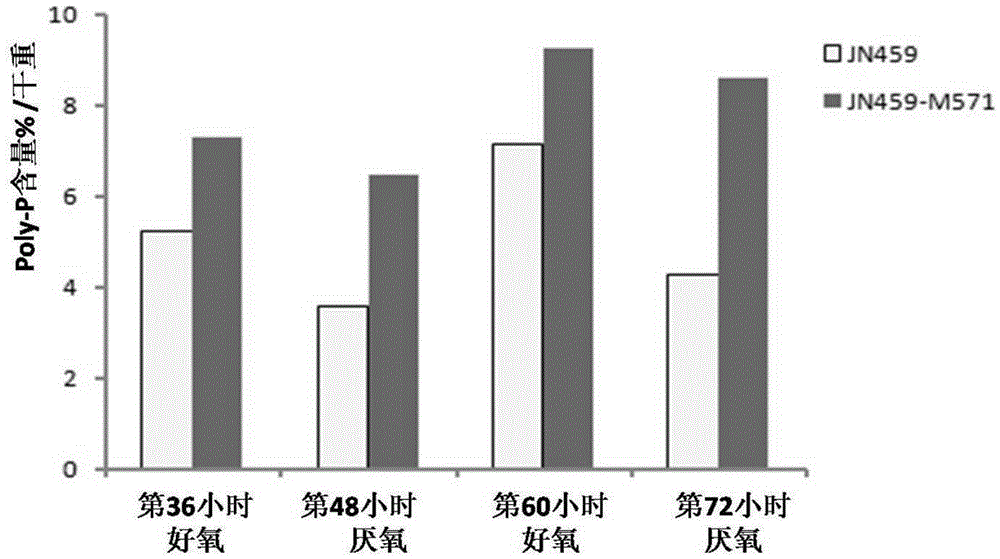
[0050] The recombinant bacterial strain constructed in Example 2 was cultivated to the logarithmic growth phase with the above-mentioned liquid medium (Example 2), and the bacterial cells were collected by centrifugation, washed with sterile water, and diluted to 10 8 Individual / ml, inoculate into new above-mentioned liquid culture medium according to 5% inoculum amount, each recombinant bacterial strain inoculates a 50ml / 250ml shaking flask. Cultivate on a shaker at 25°C and 200rpm for 48 hours, absorb 10ml of the bacterial liquid, centrifuge at 3000rpm for 10min to collect the bacterial cells, and measure the Poly-P content. Replace the remaining culture medium with sterile nitrogen to replace the air in the shaker flask, then seal the mouth of the bottle, continue to cultivate on a shaker at 25°C and 200rpm for 24 hours, absorb 10ml of bacterial solution, and centrifuge at 3000rpm for 10min to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com