Method for preparing novel genome simplified methylation sequencing library
A sequencing library and genome technology, applied in the field of genome simplified methylation sequencing library preparation, can solve the problems of difficult preparation of detection chips, lack of large-scale application, and few methylation data detection methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
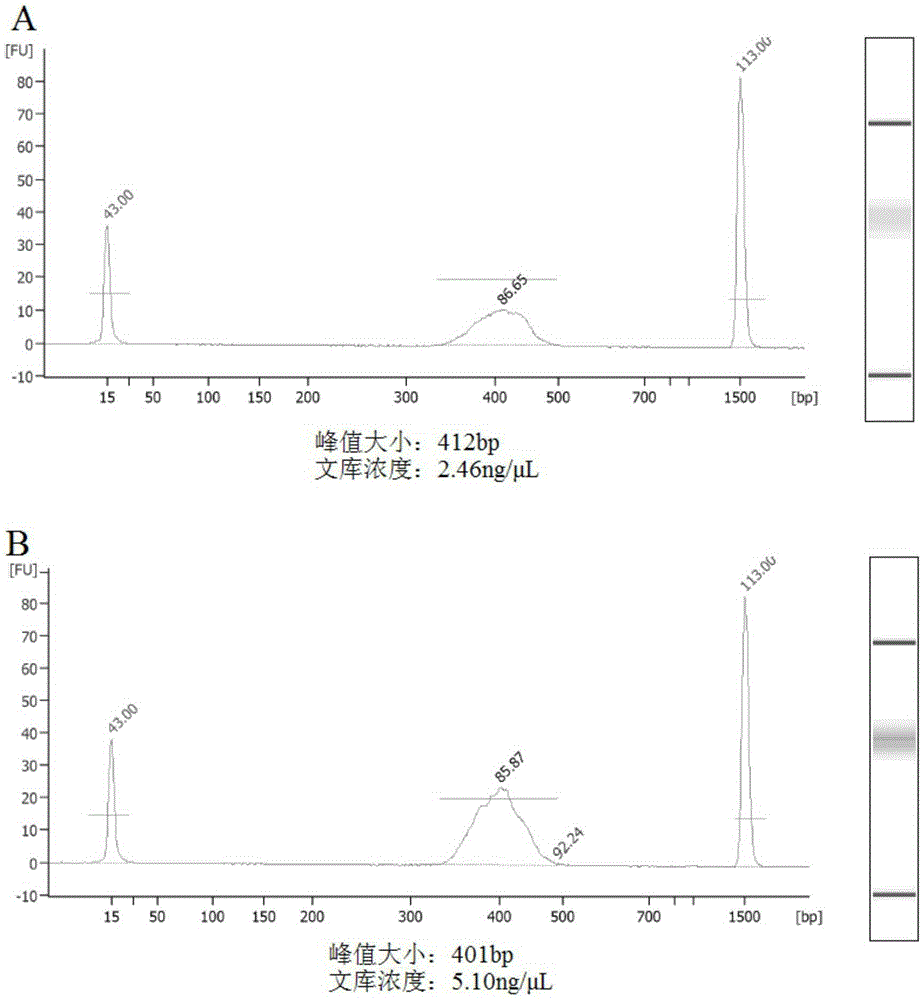
[0045] Example 1 MSAP-seq library preparation of photothermosensitive male sterile line rice Wuxiang S (WXS) and conventional rice 9311
[0046] Material background: Wuxiang S (WXS) is a light-temperature-sensitive male sterile line of rice. It is sterile under high temperature in long days and fertile in low temperature in short days. Fertility is regulated by light and temperature, which belongs to the category of epigenetics. Epigenetics is a branch of genetics that studies the heritable changes in gene expression without changing the nucleotide sequence of the gene. 9311 is an indica rice conventional rice, which is fertile under long-day high temperature and short-day low temperature, and 9311 is a positive control.
[0047] Sample processing and collection:
[0048] Table 2. Sample Handling
[0049]
[0050] 9311 two groups of materials are negative controls, which are used as background impurities; WXS two groups of materials are positive controls. The three tissu...
Embodiment 2
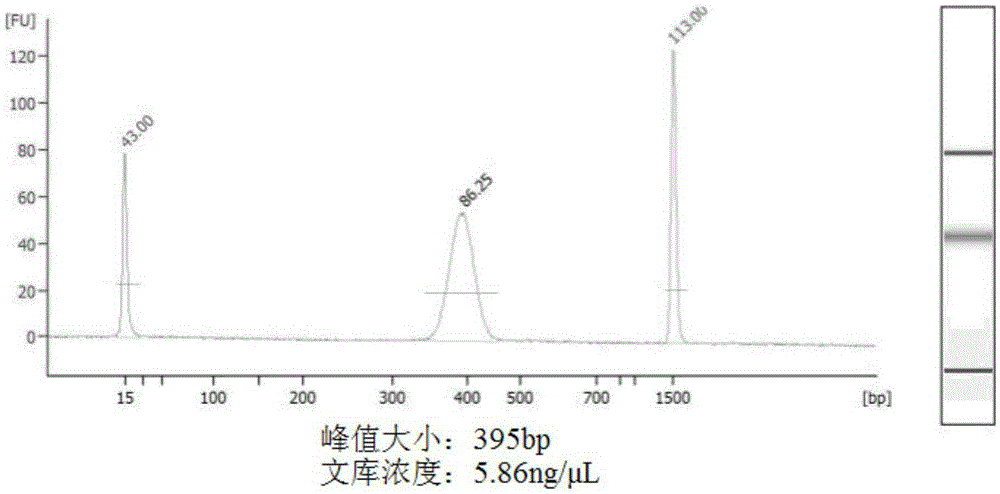
[0072] Example 2 MSAP-seq library preparation of camphor tree young leaves and mature leaves
[0073] Sample collection: Three healthy camphor trees were selected next to the School of Life Sciences of Wuhan University, three fresh leaves and three mature leaves were taken respectively, and MSAP-seq was prepared to study the methylation status changes of the same tissue at different developmental stages library.
[0074] Table 4. Barcode (Barcode) and indicator label information table in the MSAP-seq library preparation of new leaves and mature leaves of camphor tree
[0075]
[0076]
[0077] Note: Due to the small number of samples, the samples can be mixed into one pool and share the same indicator PCR primer to save the cost of library construction and sequencing.
[0078] The specific MSAP-seq library preparation process is as follows:
[0079] (1) Extraction of DNA samples. Genomic DNA was extracted from fresh and mature leaves of camphor tree with 2×CTAB extrac...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com