Method for establishing DNA (deoxyribonucleic acid) fingerprint spectra of peanut aspergillus flavus in different producing areas with ISSR (inter-simple sequence repeat) molecular marker technology
A technology of DNA fingerprinting and Aspergillus flavus, which is applied in the field of food safety, and achieves the effect of stable method, easy mastering and good primer specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1—the inventive method is stable, reproducible, easy to master
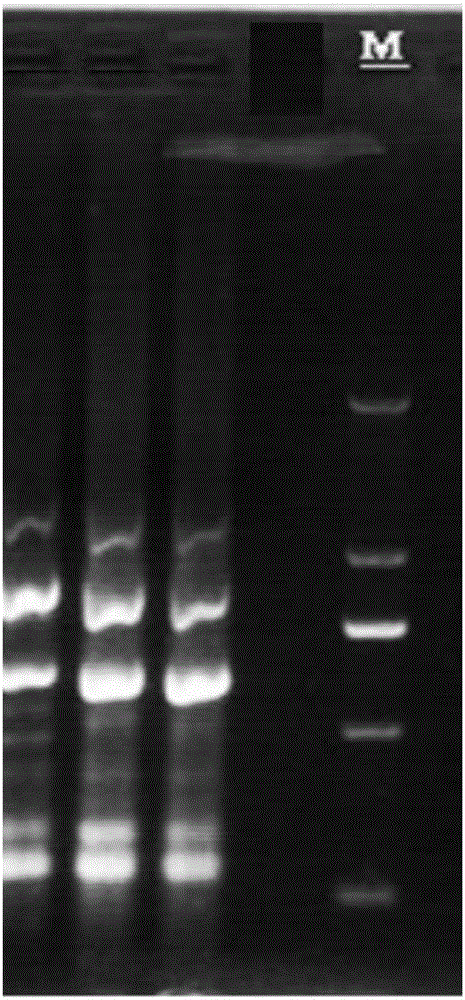
[0037] According to the method of the present invention, a strain of Aspergillus flavus is carried out three parallels of the ISSR mark of R1 primer, and its collection of figures is shown in figure 1 . Where M is a DNA marker. figure 1 It can be seen that the method of the present invention is stable, has good reproducibility, clear background and high band brightness.
Embodiment 2
[0038] Embodiment 2—The present invention can trace the source of Aspergillus flavus pollution in the main peanut production area of China to a single production area or a smaller area
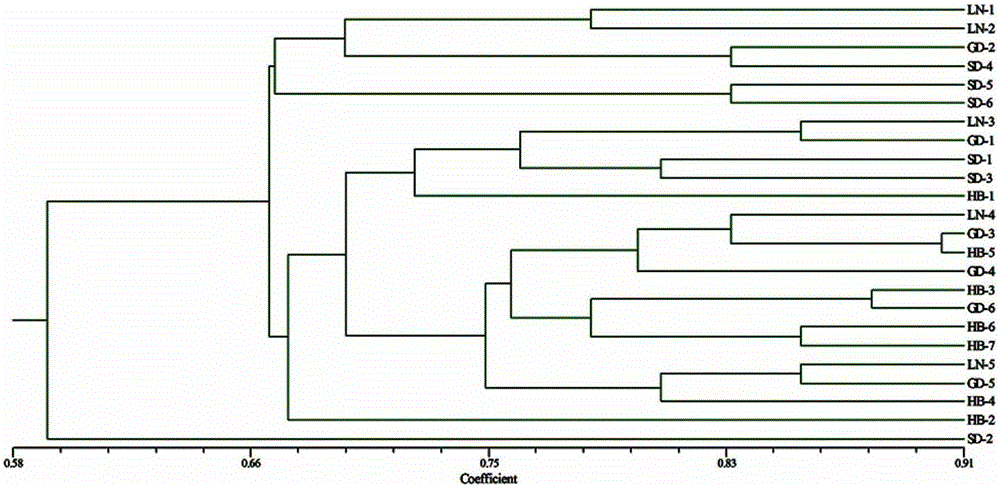
[0039] According to the method of the present invention, 24 strains of Aspergillus flavus isolated from different regions are subjected to ISSR markers of R1, R2, and R3 primers, and the results are clustered and analyzed, and the map figure 2 . From figure 2 It can be seen that the bacterial strains in each region can be separated from the bacterial strains in other regions by this method. Therefore, the present invention can trace the source of Aspergillus flavus pollution in the main peanut producing regions of China to a single producing region or a smaller area.
Embodiment 3
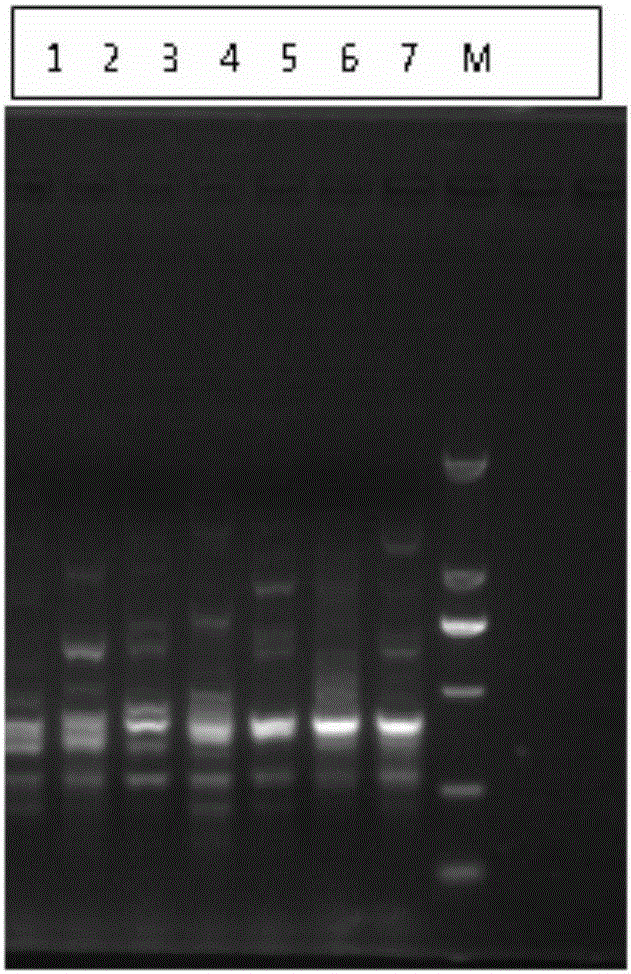
[0040] Example 3—Peanut Aspergillus flavus sample: 7 strains of Aspergillus flavus isolated from peanuts produced in Xiangyang Village, Baoshan Town, Jiaonan City, Qingdao City, Shandong Province
[0041] (1) Isolation of 7 strains of Aspergillus flavus: isolate, purify and identify Aspergillus flavus from peanuts produced in Xiangyang Village, Baoshan Town, Jiaonan City, Qingdao City, Shandong Province.
[0042] Take 10 g of peanuts from different origins, add 90 mL of 0.1% peptone sterile water (w / v), shake at room temperature for 30 minutes, and make a 10-1 bacterial suspension; take 0.1 mL of the bacterial liquid, and spread it on the DG18 medium, Incubate in the dark at 30°C for 5 days. Pick Aspergillus flavus with yellow spores and perform secondary streak isolation on DG18 medium until a single colony is obtained. Pick a single colony of Aspergillus flavus on the MEA slant test tube medium, culture it at 30°C for 3 days, and then store it at 4°C. The identification of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com