Application of AtSRT2 gene in improvement of salt resistance of plants
A plant and active technology, applied in the field of molecular genetics, can solve the problem of no AtSRT2 mediating plant salt tolerance, and achieve the effects of increased plant survival rate, enhanced salt tolerance, and good plant growth
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
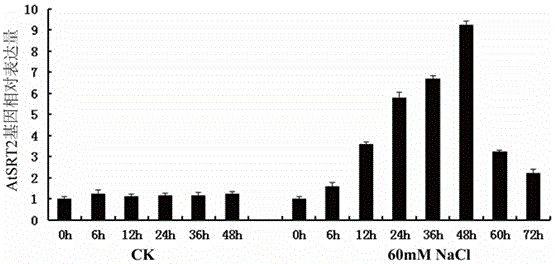
[0038] Embodiment 1, seed germination period AtSRT2 Expression induced by salt stress
[0039] 1. Extraction of Arabidopsis total RNA
[0040] The seeds of wild type Arabidopsis Columbia0 were treated with 70% (volume fraction) alcohol in an ultra-clean workbench for 3 minutes, and then washed twice with sterile water, each time for 1 minute, with 5‰ (mass volume ratio) sodium hypochlorite. After sterilizing for 10 minutes, wash twice with sterile water for one minute each time. The control group sowed sterile seeds in MS0 liquid medium with a pipette gun, and transferred them to the tissue culture room after vernalization at 4°C for 3 days. Samples were taken at 0h, 6h, 12h, 24h, 36h, and 48h. At the same time, the salt-treated group used a pipette gun to sow sterile seeds in MS0 liquid medium containing 60 mM NaCl, and after vernalization at 4°C for 3 days, they were transferred to the tissue culture room. Samples were taken at 60h and 72h.
[0041] Total RNA was extra...
Embodiment 2
[0050] Embodiment 2, protoplast subcellular localization
[0051] 1. Construction of subcellular localization vector
[0052] Specific primers AtSRT2-F2 and AtSRT2-R2 were designed according to the CDS sequence of the AtSRT2 gene provided on the TAIR website to amplify the full length of the AtSRT2 gene (remove the stop codon). Use Infusion enzyme to connect the target fragment to the BamHI-digested subcellular localization vector 16318GFP. The ligation reaction system is: 2 μl InfusionHDEnzymePremix, 5 μl 16318GFP digestion product, 3 μl PCR amplification product, and react at 50°C for 15 minutes. After the ligation reaction, heat shock transformed E. coli Top10. PCR detection of recombinant plasmids. Positive clones were sequenced. The plasmid was extracted to obtain the recombinant plasmid 116318-AtSRT2-GFP.
[0053] The primer sequences are:
[0054] 16318-AtSRT2-GFP-F: 5'-TATCTCTAGA GGATCC ATGCTTTCTATGAACATGAGAAG-3' (the underlined part is the recognition site of...
Embodiment 3
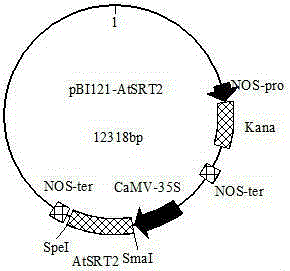
[0075] Embodiment 3, AtSRT2 Gene cloning and obtaining overexpression lines
[0076] 1. Cloning of AtSRT2 gene and construction of recombinant expression vector pBI121-AtSRT2
[0077] The wild-type Arabidopsis Columbia0 at the adult plant stage was used as the experimental material, and the total RNA was extracted by the TRIzol method. The qualified RNA was reverse-transcribed into cDNA with the reverse transcription kit of TaKaRa Company, and stored at -20°C for future use. According to the TAIR website provided AtSRT2 Design specific primers for the CDS sequence of the gene AtSRT2 -F2 and AtSRT2 -R2, amplified AtSRT2 Gene full length. The reaction system includes 2×GCBuffer 25μl, dNTPMix 4μl, upstream and downstream primers 1μl, Primerstar 0.3μl, ddH 2 O16.7μl, template cDNA 2μl (the above PCR reaction reagents are from TaKaRa company PCR amplification kit). The PCR amplification program was pre-denaturation at 94°C for 10 min, denaturation at 94°C for 30 s, anneali...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com