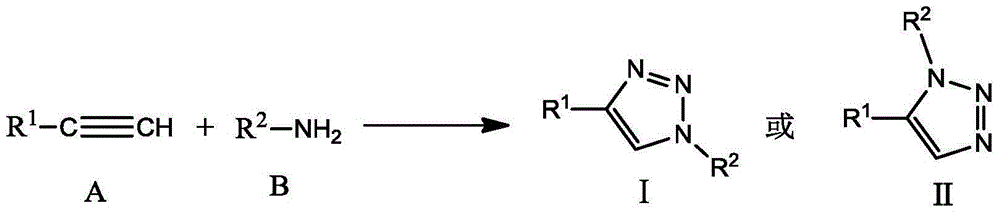
Method for marking and modifying biomacromolecules by one-pot process
A technology of biological macromolecules and small molecules, which is applied in the preparation of sugar derivatives, sugar derivatives, and sugar derivatives, and can solve problems such as perishable, highly toxic and explosive, and harsh reaction conditions.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
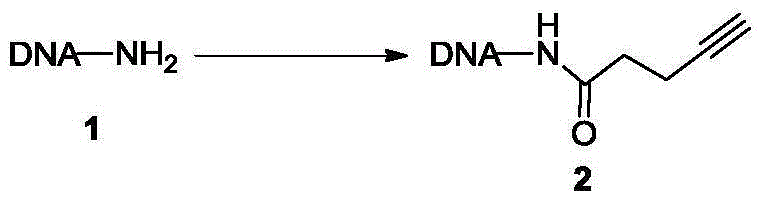
[0086] Example 1 Synthesis of alkyne-modified oligodeoxynucleotides
[0087]
[0088] Oligodeoxynucleotide 1 (molecular weight: 7743.0 Da, 100.0 nmol) modified at the 5' terminal amino group was dissolved in 20.0 μL of phosphate buffered saline solution (pH=7.4, 0.5 M) to obtain an oligodeoxynucleotide solution. 4-alkynylpentanoic acid (0.1mg, 1.0μmol), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (0.38mg, 2.0μmol) and N-hydroxysuccinate After imide (0.23 mg, 2.0 μmol) was dissolved in 40.0 μL dimethyl sulfoxide, the above oligodeoxynucleotide solution was added, mixed and reacted overnight at 50 ° C, and then 50 μL of 0.1 M trihydroxy Methylaminomethane-hydrochloric acid buffer solution (pH=9.0), 200 μL of 0.5M sodium acetate-acetic acid buffer solution (pH=4.7) and 750 μL of absolute ethanol were placed in a freezer at -20°C for two hours and allowed to settle for two hours. Centrifuge at 14,000g at 4°C for 15 minutes for sedimentation and separation, and ...
Embodiment 2
[0092]
[0093] Dissolve (pyridin-2-yl)methylamine (2.7mg, 25.0μmol) in 40.0μL methanol, then add 20.0μL imidazolesulfonyl azide aqueous solution (3.8mg, 15.0μmol), 10.0μL copper sulfate aqueous solution (2.4mg , 15.0 μmol) and 40 μL triethylamine (2.5 mg, 25 μmol) in methanol were reacted at 60° C. for 1 hour to obtain a reaction solution.
[0094] Take 10 μL of the above reaction solution (which contains 2.3 μmol of (pyridin-2-yl) methyl azide), add copper sulfate (0.1 mg, 0.63 μmol), 2.0 μL of 0.01M tris[(1-benzyl-1 -Hydro-1,2,3-triazol-4-yl)methyl]amine in dimethyl sulfoxide solution and 10 μL aqueous sodium ascorbate (0.2 mg, 1.0 μmol) and 30 μL alkyne-modified oligodeoxynucleoside Aqueous solution of acid product 2 (5.0 nmol). After reacting at 25°C for 12 hours, add 25.0 μL tris-hydrochloric acid buffer solution (pH=9.0, 0.1M), 10.0 μL 0.1M ethylenediaminetetraacetic acid aqueous solution, and 100 μL sodium acetate-acetic acid buffer solution (pH=4.7, 0.5M) and 400...
Embodiment 3
[0098]
[0099] Dissolve 2-(thiophen-2-yl)ethylamine (3.2 mg, 25.0 μmol) in 40.0 μL of methanol, then add 20.0 μL of imidazolesulfonyl azide aqueous solution (3.8 mg, 15.0 μmol), 10.0 μL of copper sulfate Aqueous solution (2.4 mg, 15.0 μmol) and 40.0 μL triethylamine (2.5 mg, 25.0 μmol) in methanol were reacted at 60° C. for 1 hour to obtain a reaction solution.
[0100] Take 10.0 μL of the above reaction solution (which contains 2.3 μmol of 2-(thiophen-2-yl)ethyl azide), add copper sulfate (0.1 mg, 0.63 μmol), 2.0 μL, 0.01M tris[(1-benzyl 1-hydrogen-1,2,3-triazol-4-yl)methyl]amine in dimethyl sulfoxide solution and 10.0 μL sodium ascorbate aqueous solution (0.2 mg, 1.0 μmol) and 30 μL alkynyl-modified oligo Aqueous solution (5.0 nmol) of polydeoxynucleotide product 2, react overnight at 25°C, add 25.0 μL tris-hydrochloric acid buffer solution (pH=9.0, 0.1M), 10.0 μL, 0.1M ethyl alcohol Diaminetetraacetic acid aqueous solution, 100 μL of sodium acetate-acetic acid buffer s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com