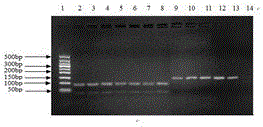
American ginseng DNA detection reagent box and identification method
A detection kit and identification method technology, applied in the field of identification of traditional Chinese medicinal materials, can solve problems such as inconvenience in practical application, difficulty in popularization and application, differences in drug efficacy and function, and achieve reliable detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] 1. Pretreatment of test samples
[0070] Test samples (two types, 1 is authentic American ginseng, 2 is ginseng, American ginseng counterfeit, both provided and identified by the China Institute for the Control of Pharmaceutical and Biological Products), each take 0.03g, rinse with pre-treatment solution after scrubbing, and dry at room temperature , irradiate with ultraviolet light for more than 30 minutes, use scissors to cut the sample to about 1-2mm, put it in a centrifuge tube, and weigh it for later use;
[0071] 2. Genomic DNA Extraction
[0072] (a) Sample lysis Add 1ml of lysate to the pretreated sample, place it in a water bath, bathe at 65°C for 30 minutes, and rotate at 100r / min;
[0073] (b) After precipitation and lysis, centrifuge the sample at 13000r / min at 4°C for 10min, absorb the supernatant, add an equal volume of precipitation solution, mix well, centrifuge at 13000r / min at 4°C for 2min, discard the supernatant, and keep the precipitate;
[0074] (c...
Embodiment 2
[0093] Example 2 Identification of commercially available American ginseng medicinal materials
[0094] 1. Materials
[0095] 5 samples of commercially available American ginseng, 1 genuine product of American ginseng (provided and identified by Jilin Drug Control Institute)
[0096] 2. Method
[0097] 2.1 Pretreatment of test samples Take 0.03g of each test sample, scrub it clean, rinse with pretreatment solution, dry at room temperature, irradiate with ultraviolet light for more than 30min, use scissors to cut the sample to about 1-2mm, and place it in a centrifuge tube , weighed for use;
[0098] 2.2 Genomic DNA extraction
[0099] (a) Sample lysis Add 1ml of lysate to the pretreated sample, place it in a water bath, bathe at 65°C for 30 minutes, and rotate at 100r / min;
[0100] (b) After precipitation and lysis, centrifuge the sample at 13000r / min at 4°C for 10min, absorb the supernatant, add an equal volume of precipitation solution, mix well, centrifuge at 13000r / m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com