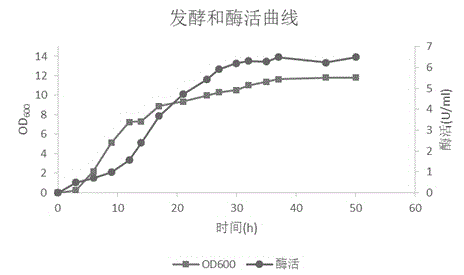
Cre/lox system based method for construction of resistance gene free chromosome integrated recombinant bacillus subtilis expressing D-psicose 3-epimerase
A Bacillus subtilis, no resistance gene technology, applied in the field of enzyme genetic engineering, can solve the problems of complex process, difficult separation and purification, and many by-products, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Embodiment 1: Construction of P43-DPE expression unit
[0024] Introduce the P43 promoter by overlapping PCR, use the primer pair P1 / P2 to amplify the P43 promoter, use the primer pair P3 / P4 to amplify the DPE enzyme gene connection, and design the primers as follows:
[0025] P1: 5’-CC GCTAGC TG ATAGGTGGTA TGTTTTC-3'
[0026] P2: 5’-TAATAAATAC CATGCTTCAT GTGTACATTC CTCTCT-3’
[0027] P3: 5’-AGAGAGGAAT GTACACATGA AGCATGGTAT TTATTA-3’
[0028] P4: 5'- GTC GAC TTAC TTCCATTCAA GCATG -3’
[0029] PCR product 1 was obtained by using P1 and P2 as primers by PCR method. PCR product 2 was obtained by using P3 and P4 as primers.
[0030] Overlap PCR was performed using PCR product 1 and PCR product 2 as templates and P1 and P4 as primers. Its reaction system is as follows:
[0031]
[0032] The reaction program was as follows: pre-denaturation at 98°C for 1 min; 30 cycles of 98°C for 10 s, 55°C for 15 s, and 72°C for 1 min; extension at 72°C for 5 min, and cooling t...
Embodiment 2
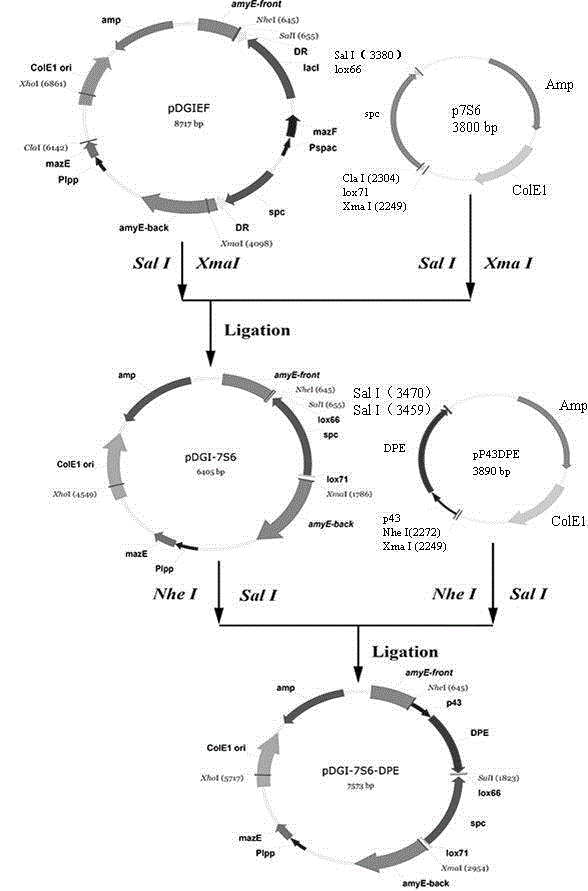
[0033] Example 2: Construction of integration vector pDGI-7S6-P43-DPE
[0034]Plasmids p7S6 and pDGIEF were digested with SalI and XmaI, respectively, and recovered lox 71- spc - lox 66 and the vector pDGIEF gene fragment were ligated overnight with T4 ligase and transformed into E. coli DH5α competent cells, spread to LB solid medium (containing 100 μg / mL spc ), cultured overnight at 37°C. Pick positive clones, extract plasmids, and perform double enzyme digestion verification. The verified correct plasmid was named pDGI-7S6.
[0035] Plasmids pDGI-7S6 and pP43-DPE were digested with NheI and SalI respectively, and the gene fragments pDGI-7S6 and P43-DPE were recovered, ligated with T4 ligase overnight, and then transformed into E. coli DH5α competent cells, coated with s pc (100 μg / mL) LB plates, cultured overnight at 37°C. Pick positive clones, extract plasmids, perform double enzyme digestion and sequencing verification. The verified and sequenced correct p...
Embodiment 3
[0036] Example 3: Chromosomal Integration
[0037] The recombinant plasmid pDGI-7S6-P43-DPE was linearized with XhoI and then transformed into the host bacteria B. subtilis In 1A751 competent medium, apply to LB solid medium (containing 100 μg / mL spc ), cultured overnight at 37°C, and the transformants of positive clones were selected as B. subtilis 1A751-DPE-spc ( figure 2 ). Making recombinant bacteria B. subtilis 1A751-DPE-spc competent, transform the pTSC plasmid with Cre gene into competent, use the Em (250 μg / mL) LB plate screening, pick positive clones, inoculate with toothpick containing Em (250 μg / mL) and Spc (100 μg / mL) double resistance plate and containing only Em (250 μg / mL) resistance plate, select only with Em resistant without Spc For resistant positive clones, increase the temperature and lose the plasmid pTSC to obtain recombinant bacteria B. subtilis 1A751 / lox -DPE.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com