Method for confirming family-specific genetic disease-associated allelic haplotype variant signatures
An allele and haplotype technology, applied in the fields of genetics and molecular biology, can solve the problem of no unified standardization method, and achieve the effect of easy operation and understanding, low sequencing cost, and saving detection cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
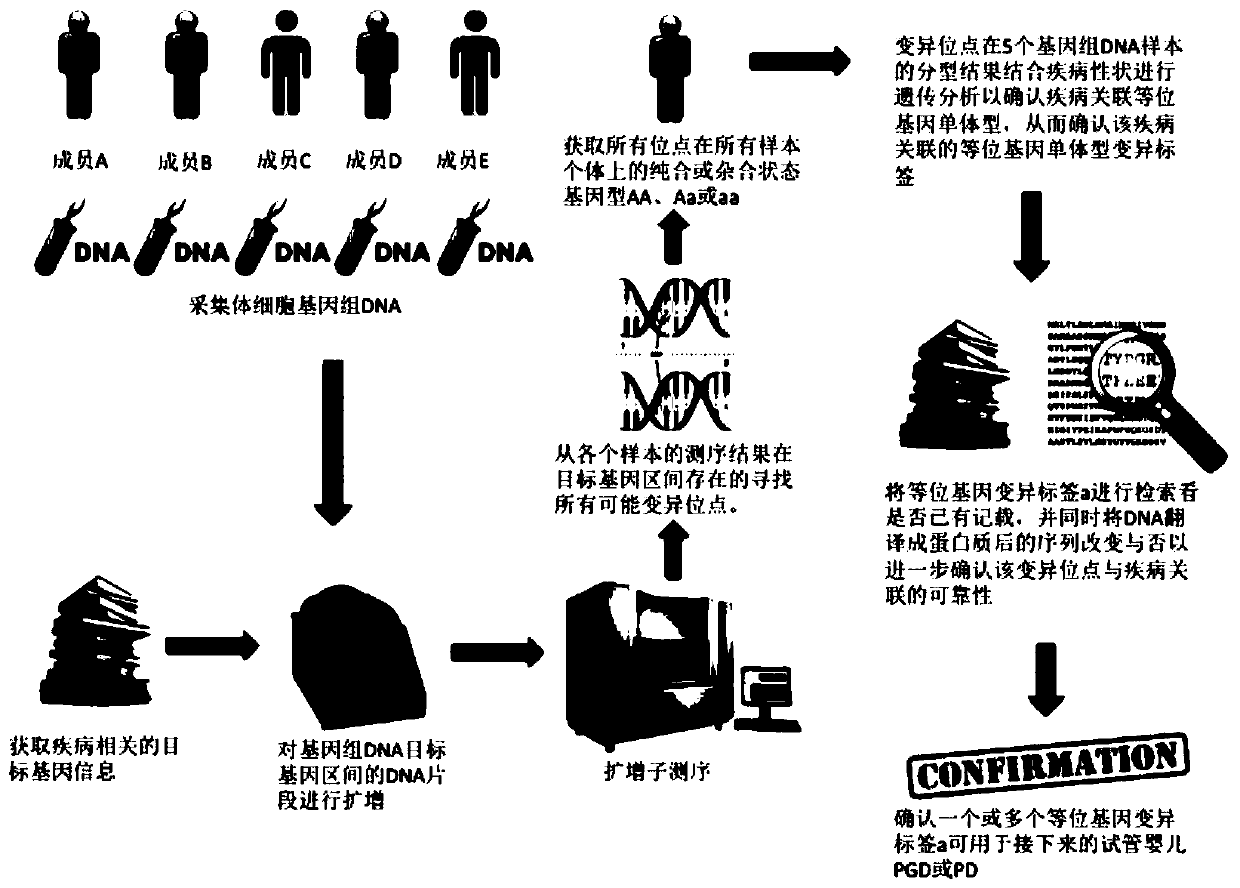
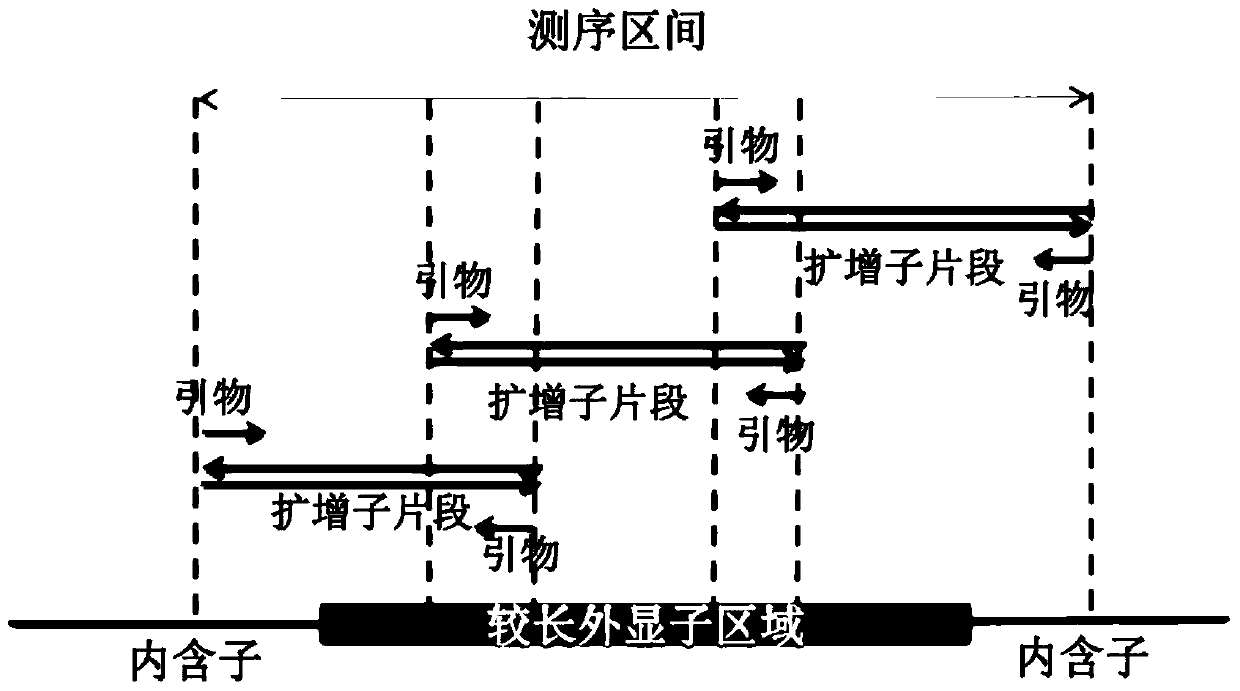
[0038] For a method for confirming the allelic haplotype variant signature associated with Hereditary Multiple Exostoses in a family, see figure 1 , including the following steps,
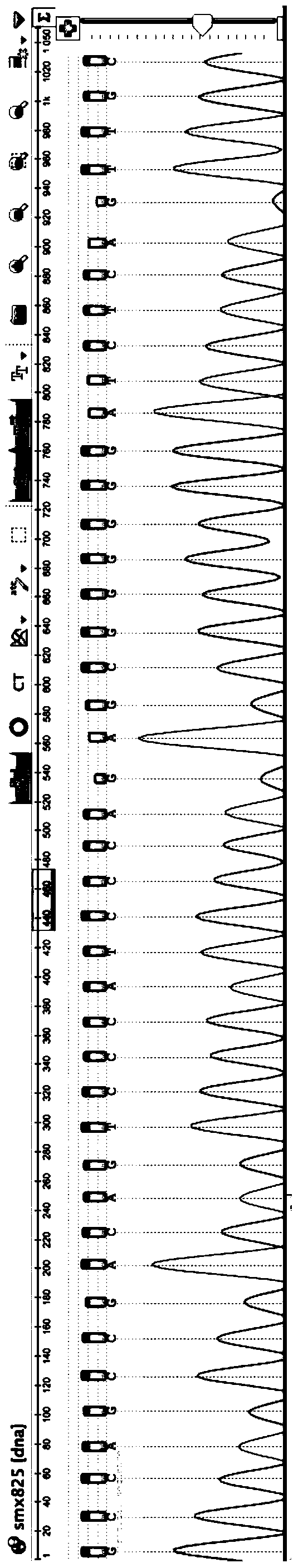
[0039] 1) Select 5 people from the Mendelian genetic disease family to extract genomic DNA samples. Select 5 family members, member A is about to give birth and suffers from a family Mendelian genetic disease; member B is the father of member A; member C is the mother of member A; member D is the cousin of member A, and is the disease family The positive control of member E; member E is the cousin of member A, and is the negative control of the disease family. Among them, member D and member E were from paternal family members at risk of disease. Collect 5ml of peripheral venous blood from the above-mentioned 5 members respectively, and use TaKaRaMiniBEST Whole Blood DNA Extraction Kit to extract DNA. For details, please refer to the operation manual of the kit. Five members extracted genomic DN...
Embodiment 2
[0055] Search allelic haplotype variation tags found in Example 1, confirm the records, and translate the DNA into protein. If the amino acid sequence of the protein changes, it will help to further confirm the variation of the allele at this site. Reliability of disease associations. We further conducted a literature survey on the deletion mutation and found that the deletion mutation had been recorded in the literature (Seki H, Kubota T, Ikegawa S, Haga N, Fujioka F, Ohzeki S, etal. Mutation frequencies of EXT1 and EXT2in43 Japanese families with hereditary multiple exostoses. American journal of medical genetics. 2001;99(1):59-62. Epub2001 / 02 / 15. PubMed PMID: 11170095.). Thus, it was determined that the deletion variant could be used as a confirmatory allelic haplotype signature. If there is no literature record after searching, it means that the variant site has not been found yet, and it belongs to a new family genetic variation label, which is worthy of further research...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com