Novel biomarker for evaluating lung cancer disease through plasma
A marker, plasma technology, applied in measurement devices, instruments, scientific instruments, etc., can solve problems such as affecting the application of polyamines and failing to find specific markers for lung cancer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0014] Example 1 Lung cancer and healthy human plasma sample collection method
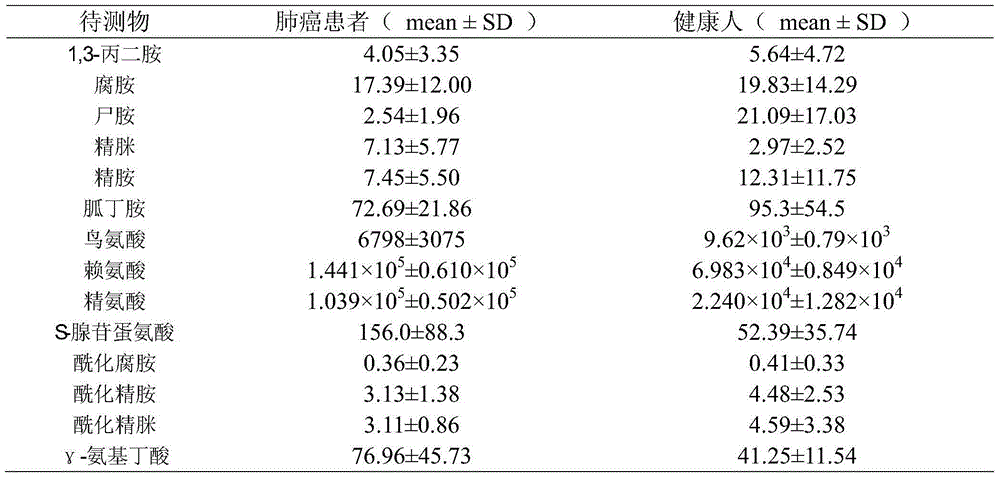
[0015] Collect 50 patients with lung cancer confirmed by pathological cytology, including 30 males and 20 females, aged 27-77 years, with an average of 53 years old. The control group consisted of 50 cases, all of whom were healthy adults, confirmed by blood and urine routine tests, including 30 males and 20 females, aged 27-65 years, with an average of 52 years old. Collect venous blood from lung cancer patients and healthy people. Immediately after collection, the blood samples should be centrifuged at 3500rpm for 10 minutes to separate the plasma. The separated plasma samples should be placed in labeled test tubes and stored in an ultra-low temperature refrigerator below -80°C.
[0016] Take 250 μL of plasma sample in EP tube, add 50 μL of methanol:water (20:80, v:v) mixed solution and 50 μL of solution internal standard solution (1,6-hexamethylenediamine, 100 ng / mL), vortex 30s. After adding...
Embodiment 2
[0017] Example 2 Detection method of polyamine samples in lung cancer and healthy human plasma
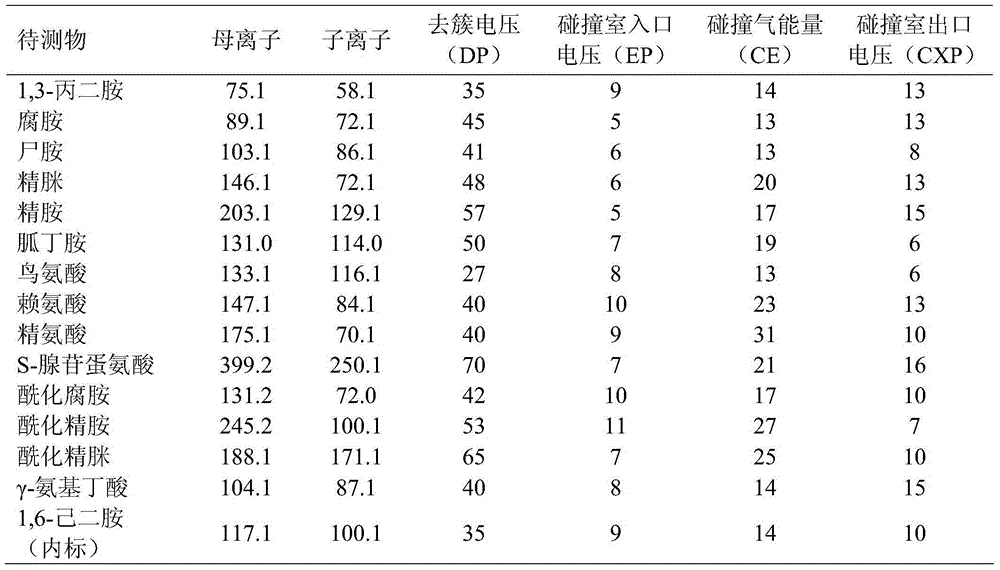
[0018] The 5 μL supernatant obtained in Example 1 was analyzed by UHPLC-MS / MS, and the detection conditions were as follows:
[0019] (1) Reagents
[0020] Standard substances 1,3-propanediamine, putrescine, cadaverine hydrochloride, spermidine hydrochloride, spermine hydrochloride, agmatine sulfate, acylated putrescine hydrochloride, acylated spermidine salt Salt, acylated spermine hydrochloride, ornithine hydrochloride, lysine, arginine, S-adenosylmethionine, and heptafluorobutyric acid are all products of Sigma. HPLC grade methanol was the product of Fisher Company, other chemical reagents were only of analytical grade, and water was deionized redistilled water.
[0021] (2) Instruments
[0022] Prominence UFLC XR ultra-fast high-separation liquid chromatograph equipped with LC-20AD infusion pump, SIL-20AC autosampler, CTO-20AC column oven and DGU-20A3 degasser (Shimadzu Corp...
Embodiment 3
[0031] Example 3 Mathematical statistics method for lung cancer samples
[0032] The data that embodiment 2 records is first carried out outlier test, adopts Hampel test method to analyze, and its concrete steps are:
[0033] 1. Calculate the mean (M e ), which divides this data set into high and low parts.
[0034] 2. Calculate r based on the mean of all elements in the data set i
[0035] r i =(x i -M e ) is the simple data in the data set,
[0036] x belongs to the data set 1--n, n is the number of elements in the data set, M e is the mean
[0037] Calculate the deviation M e|ri| The mean value of , according to the condition|r i |≥4.5M e|ri|
[0038] When the above conditions are true, the values in the data set are considered outliers.
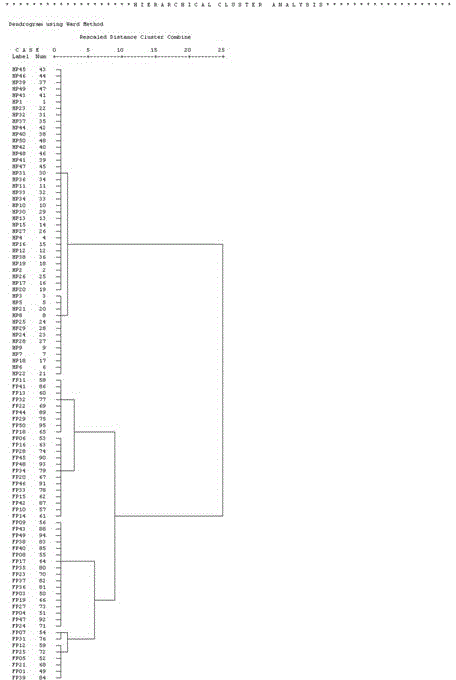
[0039] The data of 3 cases of lung cancer and 2 cases of healthy people were removed. Then, the data of lung cancer plasma and healthy human plasma were assigned 1 and 2 as dependent variables, and 14 polyamine analytes we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com