Esterase as well as encoding genes and application thereof
A technology of esterase and gene, which is applied in the field of esterase and its coding gene and application, and can solve problems such as the inability to obtain S-type products
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
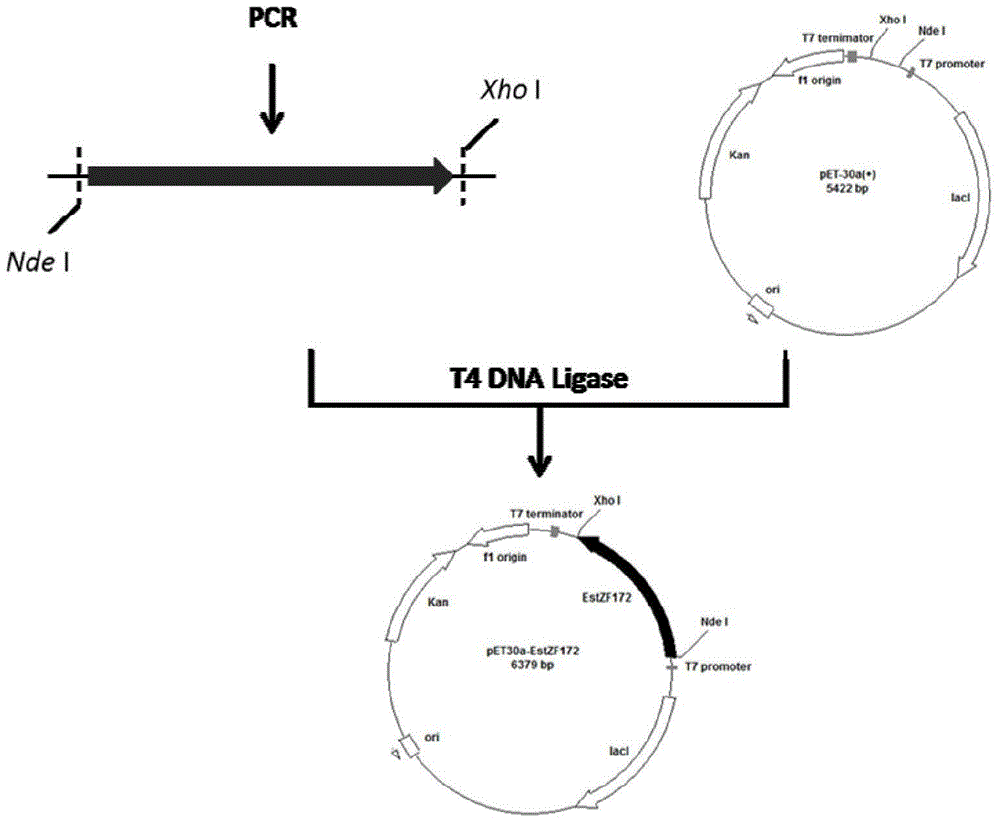
[0053] The construction of the recombinant plasmid of embodiment 1 esterase gene
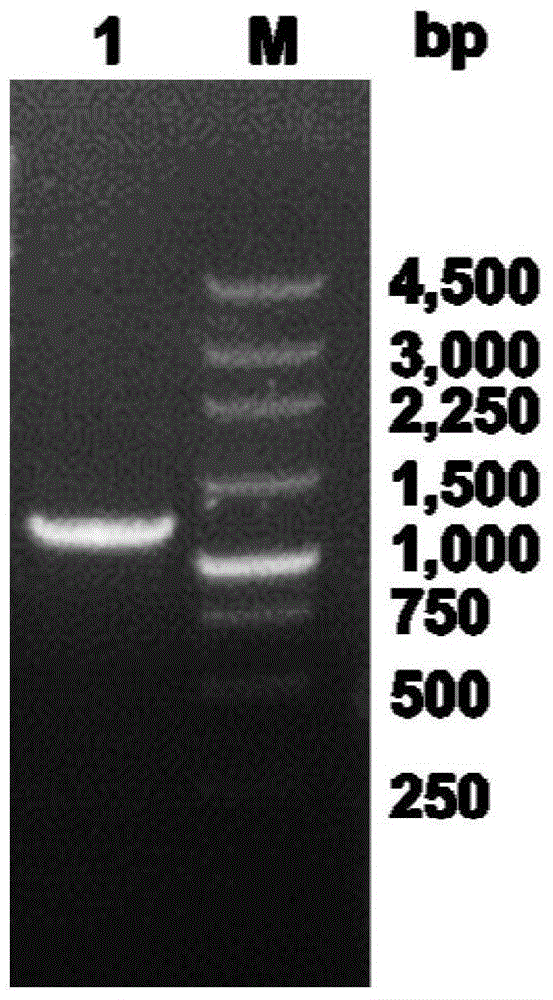
[0054] (1) Cloning of esterase gene
[0055] According to the genome DNA sequence of Pseudomonas (Pseudomonas) CGMCC NO.4184, a pair of primers were designed, and the primer sequences were as follows:
[0056] The upstream primer is: 5'-GGGAATTC CATATG CAGATCCAGGGTCACTATGAGCTGAAGTTCG-3', (as shown in SEQ ID NO.3), wherein the underlined part is the Nde I restriction site;
[0057] The downstream primer is: 5'-CCC CTCGAG AAGGCAAGTGCCAAGAACGCGTACCA-3', (as shown in SEQ ID NO.4), wherein the underlined part is the Xho I restriction site.
[0058] Using Pseudomonas sp. (Pseudomonas sp.) CGMCC NO.4184 genomic DNA as a template for PCR amplification, the PCR reaction system and reaction conditions are as follows:
[0059] PCR amplification system:
[0060]
[0061] PCR amplification conditions:
[0062] 1) Pre-denaturation: 95°C for 5 minutes;
[0063] 2) Denaturation: 98°C for 10s; Anneal...
Embodiment 2
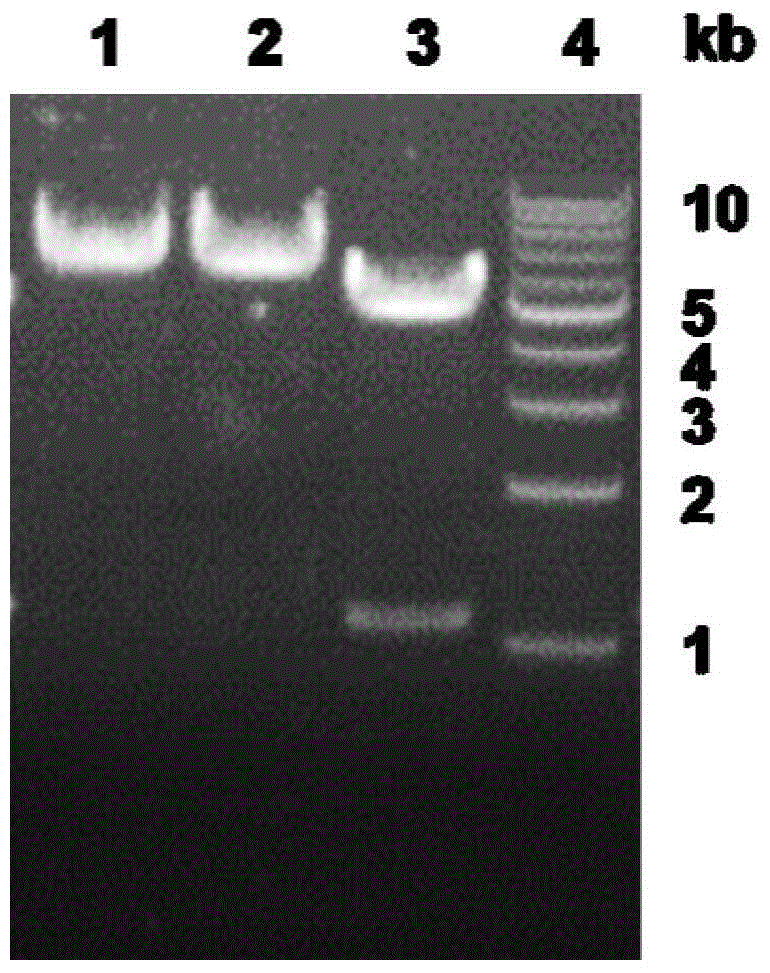
[0072] Example 2 Construction of recombinant E.coli BL21(DE3) to induce expression of esterase
[0073] The recombinant cells carrying the pET30a-estZF172 plasmid were inoculated in 5 mL LB liquid medium (containing 50 g / mL kanamycin), and cultured overnight at 37° C. and 200 rpm. The above culture was inoculated in 50mL LB liquid medium (containing 50g / mL kanamycin) at a ratio of 1%, and cultivated to OD at 37°C and 200rpm. 600 When reaching 0.6-0.8, add IPTG to a final concentration of 0.5mmol / L. The culture was induced at 18°C and 200 rpm. After 12 hours, the cells were collected by centrifugation at 12,000×g, washed twice with pre-cooled Tris-HCl buffer (300 mM, pH 8.5), and the obtained wet cells were stored at 4°C.
Embodiment 3
[0075] Resuspend the corresponding wet cells with 14mL Tris-HCl buffer (300mM, pH 8.5) at a ratio of 0.085‰ (0.085g dry cells / L), divide into 7 parts, each 2mL, add the substrate rac-2-carboxylate Ethyl-3-cyano-5-methylhexanoate such that ethyl rac-2-carboxyethyl-3-cyano-5-methylhexanoate relative to rac-2-carboxyethyl-3 -The mass concentration of ethyl cyano-5-methylhexanoate and the total amount of cell suspension was 5g / L. Put the 7 parts of the solution into a constant temperature shaker with a temperature of 20°C, 30°C, 35°C, 40°C, 45°C, 50°C and 55°C, respectively, at a speed of 200 rpm. After 1 hour, the gas phase detection method was used to detect the substrate and The concentration of the product, its retention time on the gas phase as Figure 5 As shown, peak 1 is 2-carboxyethyl-(R)-3-cyano-5-methylhexanoic acid decarboxylation (11.46min); peak 2 is 2-carboxyethyl-(S)-3-cyano Base-5-methylhexanoic acid decarboxylate (12.53min); peak 3 is 2-carboxyethyl-(R)-3-cyano...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com