Genome editing method using attachment carrier for encoding targeted endonuclease and kit
An attachment vector, genome editing technology, applied in the field of genetic engineering, can solve the problems of reducing editing efficiency and affecting editing efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
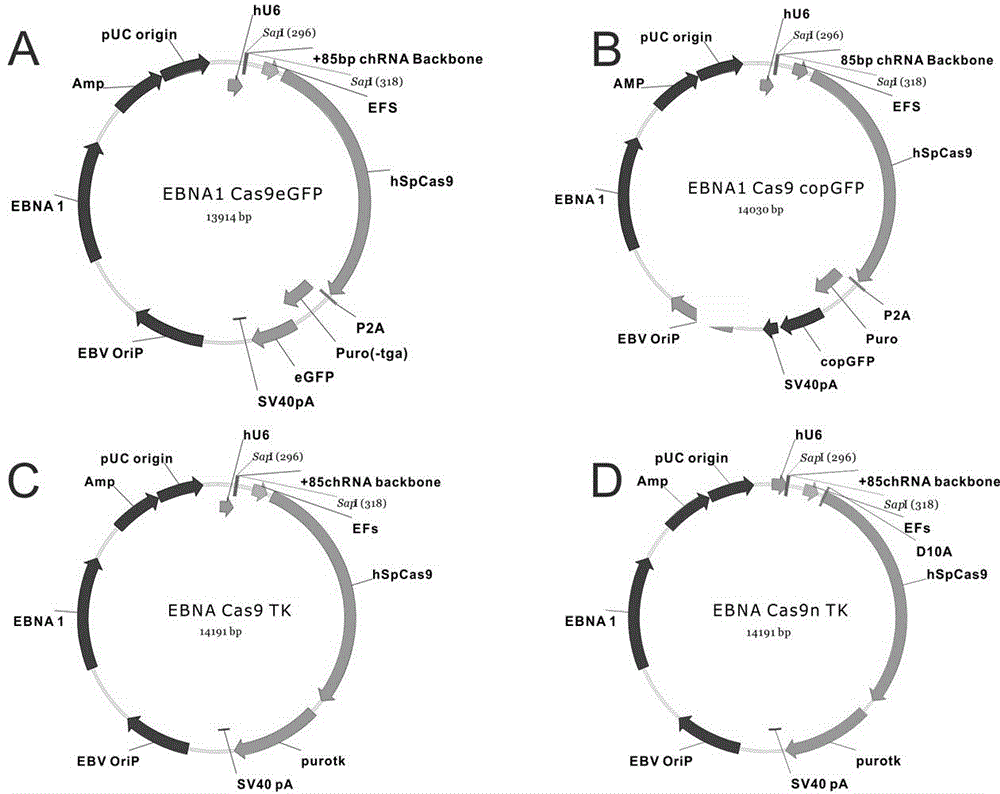
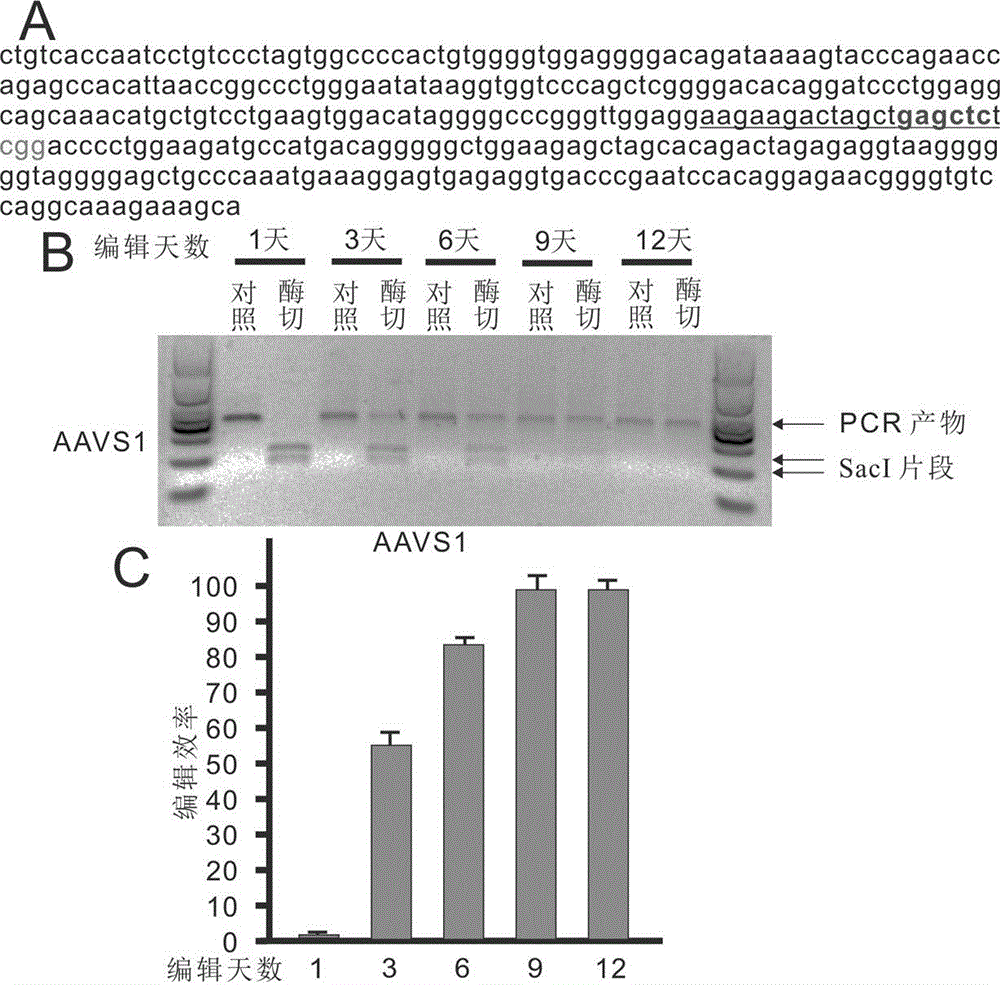
[0070] Example 1: Editing of the AAVS1 locus using the EBNA1 Cas9eGFP attachment vector
[0071] The following example details the editing of the AAVS1 locus using the EBNA1 Cas9eGFP attachment vector, the sequence of the target site is 5'- AAGAAGACTAGCTGAGCTCTCGG -3' (SEQ.ID.NO.12), and the final CGG sequence is necessary for CRISPR / Cas9 editing The PAM sequence. 1 μg for 200,000 human HEK293 cells figure 1 A Transfection of the EBNA1 Cas9eGFP plasmid. After one day of incubation, selection was performed with 2 mg / ml puromycin. On the 1st, 3rd, 6th, 9th and 12th day, DNA was extracted to identify the efficiency of digestion. There is a SacI restriction site (GAGCTC) where CRISPR / Cas9 edits, which has been marked in green ( figure 2 A). After editing occurs, this site is destroyed, and the PCR product cannot be cut by SacI, so the efficiency can be identified by enzyme digestion. Depend on figure 2 B It can be seen that editing did not occur on the first day of trans...
Embodiment 2
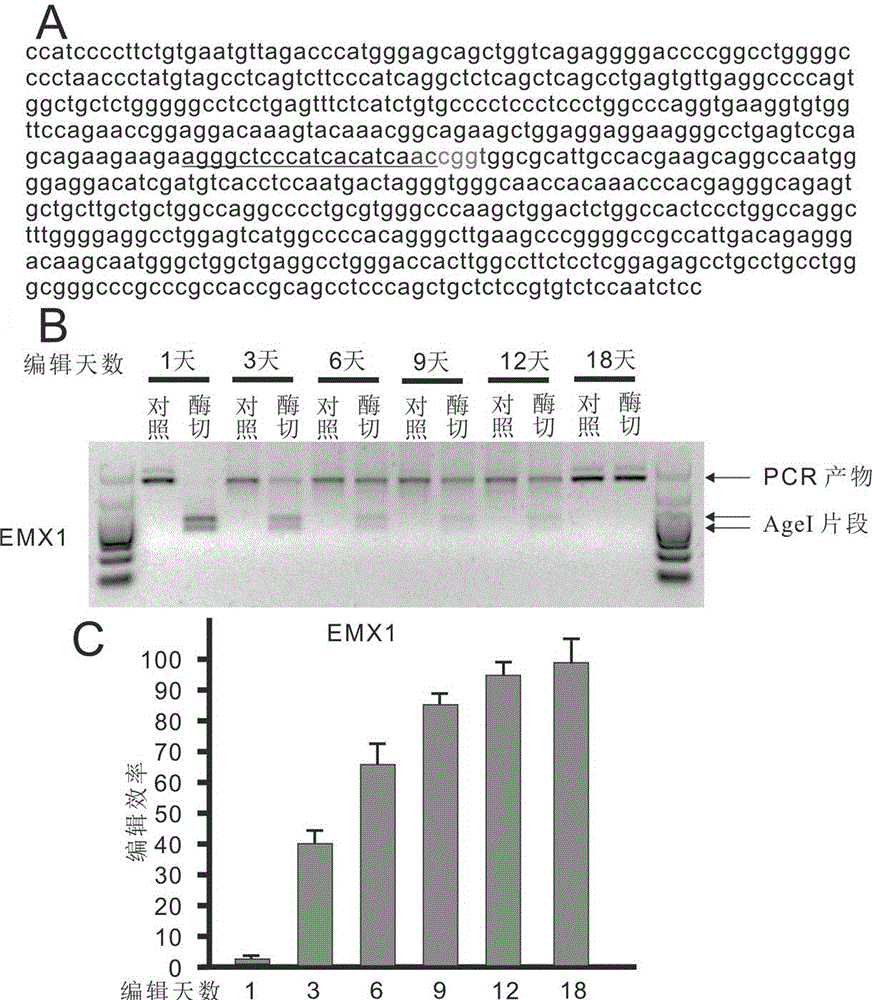
[0072] Example 2: Editing of the EMX1 locus using the EBNA1 Cas9copGFP episomal vector
[0073] The following examples detail editing of the EMX1 locus using the EBNA1 Cas9copGFP episomal vector. use as figure 1 The EBNA1 Cas9copGFP attachment vector shown in B expresses the guide RNA for EMX1. The sequence of the target site is 5'- AGGGCTCCCATCACATCAACCGG -3' (SEQ.ID.NO.13), and the last CGG sequence is the PAM sequence necessary for CRISPR / Cas9 editing. 1 μg for 200,000 human HEK293 cells figure 1 A. The plasmid transfection. After one day of incubation, selection was performed with 2 mg / ml puromycin. On the 1st, 3rd, 6th, 9th, 12th, and 18th days, DNA was extracted to identify the enzyme digestion efficiency. There is an AgeI restriction site (ACCGGT) where CRISPR / Cas9 edits, which has been marked in green ( image 3 A). After editing occurs, this site is destroyed, and the PCR product cannot be cut by AgeI, so the efficiency can be identified by enzyme digestion. ...
Embodiment 3
[0074] Example 3: Editing of the GRIN2B locus using the EBNA1 Cas9TK episomal vector
[0075] The following examples detail editing of the GRIN2B gene using the EBNA1 Cas9TK episomal vector. use as figure 1 The EBNA1 Cas9TK episome vector shown in C expresses the guide RNA of GRIN2B. The sequence of the target site is 5'-TTCCGACGAGGTGGCCATCAAGG -3' (SEQ.ID.NO.14), and the last AGG sequence is the PAM sequence necessary for CRISPR / Cas9 editing. 1 μg for 200,000 human HEK293 cells figure 1 A. The plasmid transfection. After one day of incubation, selection was performed with 2 mg / ml puromycin. On the 1st, 3rd, 6th, 9th, 12th, 18th and 22nd days, DNA was extracted to identify the enzyme digestion efficiency. There is an MSCI restriction site (TGGCCA) where CRISPR / Cas9 edits, which has been marked in green ( Figure 4 A). After editing, this site is destroyed, and the PCR product cannot be cut by MSCI, so the efficiency can be identified by enzyme digestion. Depend on ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com