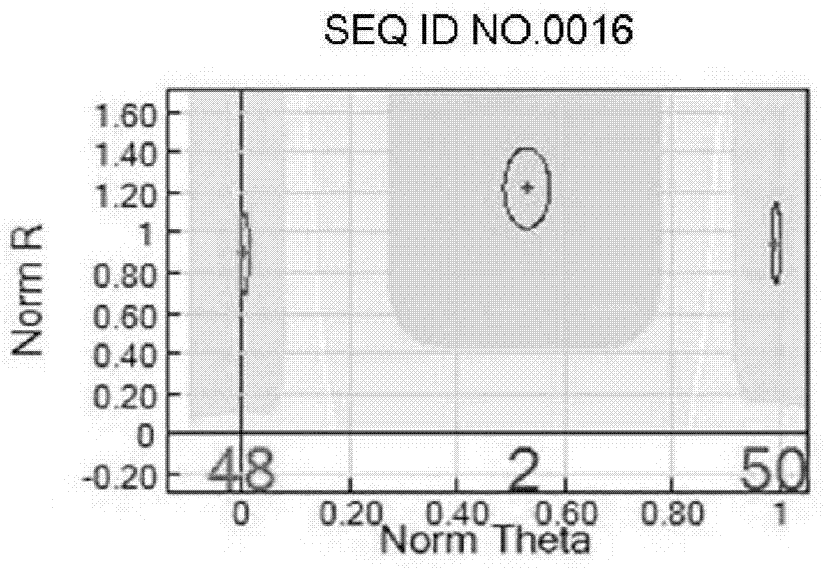
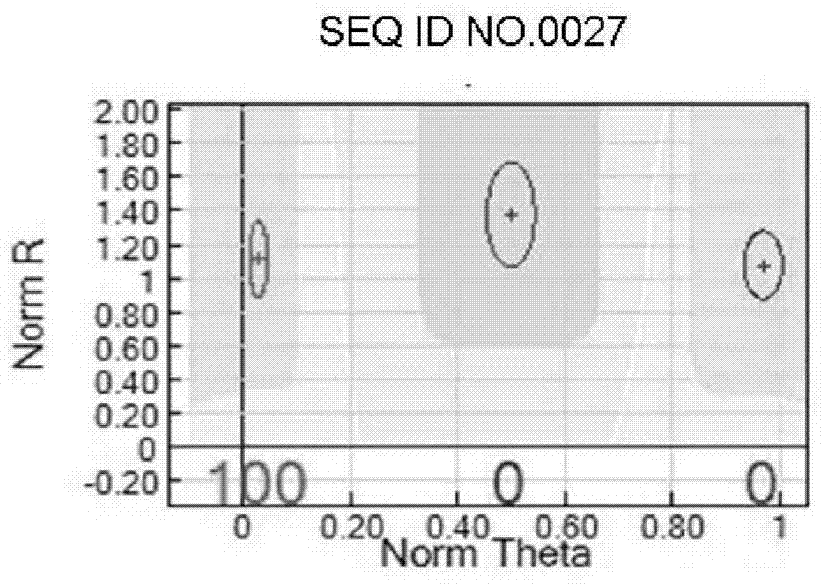
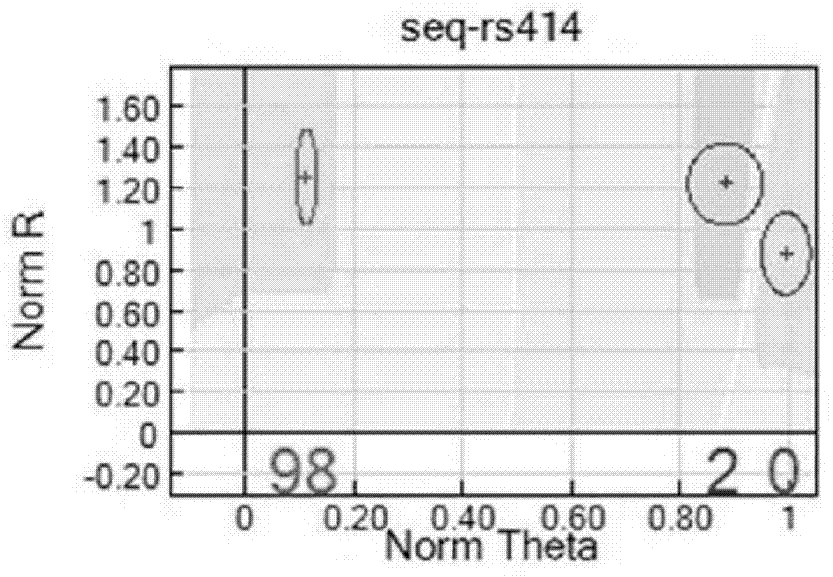
SNP chip used for identifying rice variety, preparation method and application
A variety identification and rice technology, applied in the application field of rice variety identification, can solve the problems of unsuitable variety rapid identification work, difficult data integration and sharing, low throughput, etc., to shorten the detection time, improve detection accuracy, improve The effect of detection level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: Preparation of RiceID3072 chip.
[0030] 800,000 SNP molecular markers were obtained based on the resequencing data of 950 rice cultivars (see website www.ncgr.ac.cn / ricehap2); bioinformatics software (Haploview) was used to screen marker SNPs according to linkage disequilibrium ( tagsnp) to obtain 10,000 SNP molecular markers; then further screening and evaluation is carried out for each site according to the following characteristics: (1) Whether the site evaluation score is above 0.6; (2) Whether the repeatability is above 99%; (3) Whether the deletion rate is lower than 3%; (4) Whether it is a single copy; (5) Not in transposons, repeat regions, etc.; (6) There are no other SNPs within 60 bp above and below the site; (7) The genome is evenly distributed. Finally, 3072 molecular markers were screened, and probes were designed according to the requirements of Illumina Infinium iSelectHD to make chips. The sequence listing of the 3072 SNP molecular markers ...
Embodiment 2
[0105] Example 2: Use of RiceID3072 chip
[0106] 1. DNA extraction
[0107] The seeds of 100 representative rice varieties were selected as the experimental materials, as shown in Table 1, and the plant genome extraction kit of QIAGEN was used to extract according to the standard procedure.
[0108] 2. DNA quality detection
[0109] Use a nucleic acid concentration analyzer and agarose electrophoresis to test the quality of the extracted DNA respectively to ensure that the extracted genomic DNA meets the relevant quality requirements, that is, the agarose electrophoresis shows a single DNA band without obvious dispersion; the nucleic acid concentration detector detects A260 / 280 between 1.8-2.0; A260 / 230 between 1.8-2.0; DNA concentration > 50ng / μl; chip detection DNA amount 200ng / sample. Dilute the DNA concentration to a working concentration of 50ng / / μl.
[0110] 3. Chip detection
[0111] Follow the Illumina Infinium gene chip detection standard procedure (Infinium HD ...
Embodiment 3
[0117] Example 3: Application of RiceID3072 in Identification of Rice Varieties
[0118] 1. Determination of the criteria for rice variety identification by RiceID3072.
[0119] Use RiceID3072 to detect the genotypes of the 3072 SNP molecular markers between the 100 rice varieties in Example 2, and obtain the 3072 gene bands respectively possessed by the 100 rice varieties. Any one of them is the test sample, and the other is the control sample, compare all the gene bands of the test sample and the control sample, and obtain the number of gene bands shared by the test sample and the control sample.
[0120] Utilize formula (1) to calculate the genetic similarity between test sample and control sample:
[0121] Genetic similarity GS = 2Nij / (Ni+Nj) formula (1) (Nei1979)
[0122] Among them, Nij is the number of bands shared by the tested variety and the control sample, Ni is the number of bands of the tested variety, and Nj is the number of bands of the control variety. Illus...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com