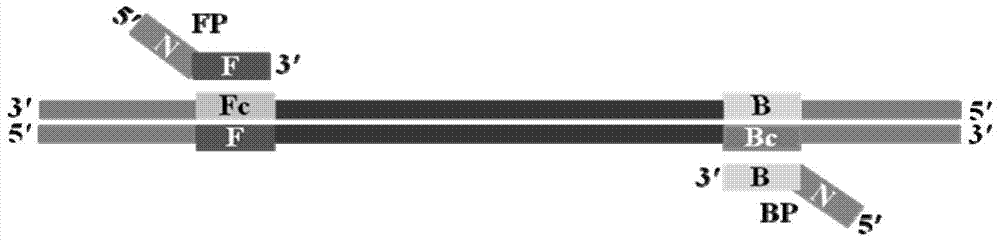Nucleic acid isothermal amplification method and application thereof by polymerase spiral reaction
A technology of constant temperature amplification and polymerase, applied in biochemical equipment and methods, microbial measurement/testing, DNA preparation, etc., can solve the problems of difficult recovery and sequencing of LAMP products, unclonable LAMP products, complex LAMP products, etc., and achieve Suitable for large-scale promotion and application, shortened reaction time, and simple primer design
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Embodiment 1, use polymerase spiral reaction (PSR) to detect whether to contain NDM-1 gene in the sample to be tested
[0088] Taking the NDM-1 gene as an example, the polymerase helix reaction (PSR) is used to detect whether the target gene (DNA) is contained in the sample to be tested. The specific method includes the following steps:
[0089] 1. Design of PSR primers
[0090] The nucleotide sequence of the NDM-1 gene (GenBank number: FN396876) was retrieved from the American gene database GenBank, and homology analysis was performed by BLAST software to find the conserved target sequence (sequence 1 in the sequence table), and then based on its conserved target sequence Design PSR primers.
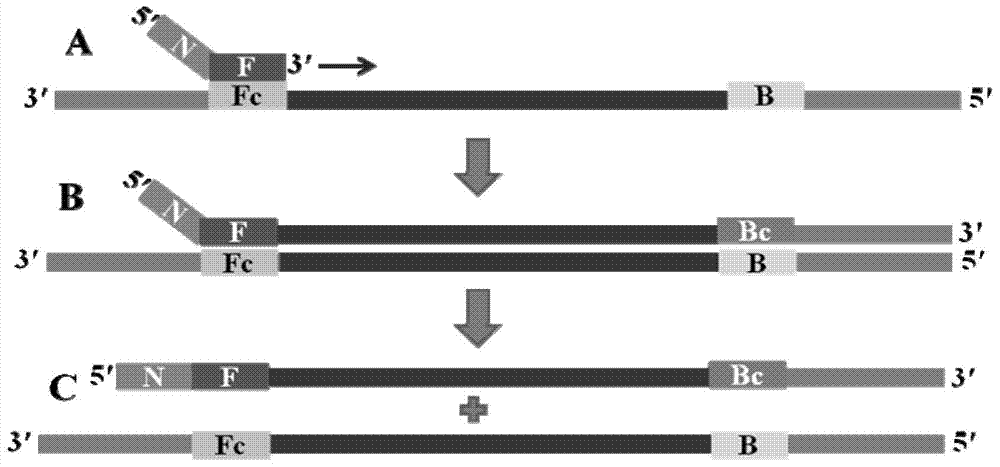
[0091] Same as ordinary PCR, first design a pair of primers F and B, F primer: 5'-GGTCGATACCGCCTGGAC-3', B primer: 5'-GCATGCAGCGCGTCCA-3', and then add a general foreign sequence at the 5' end of the two N, F primers add sequence N (5'-3': ACGATTCGTACATAGAAGTATAG) to form prime...
Embodiment 2
[0110] Embodiment 2, detect whether contain RNA in the sample to be tested with polymerase spiral reaction (PSR)
[0111] Taking the detection of H1N1 virus as an example, polymerase spiral reaction (PSR) is used to detect whether RNA is contained in the sample to be tested. The specific method includes the following steps:
[0112] 1. Design of PSR primers
[0113] The nucleotide sequence of the H1N1 gene (GenBank number: KM361419.1) was retrieved from the American gene database GenBank, and the homology analysis was performed by BLAST software to find the conserved target sequence (sequence 4 in the sequence table), and then according to its conserved target sequence Design PSR primers.
[0114] Same as ordinary PCR, first design a pair of primers F and B, F primer: 5'-GCAATGAGAACTATTGGGACTC-3', B primer: 5'-ATTTGCTGCAATGACGAGAG-3', and then add a general foreign sequence at the 5' end of the two N, F primers add sequence N (5'-3': ACGATTCGTACATAGAAGTATAG) to form primer F...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com