Method for screening and identifying genes controlled by miRNAs
A gene and genome technology, applied in the field of screening and identification of genes regulated by miRNAs, can solve the problems of heavy workload, high false positive rate, and difficulty in finding miRNAs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
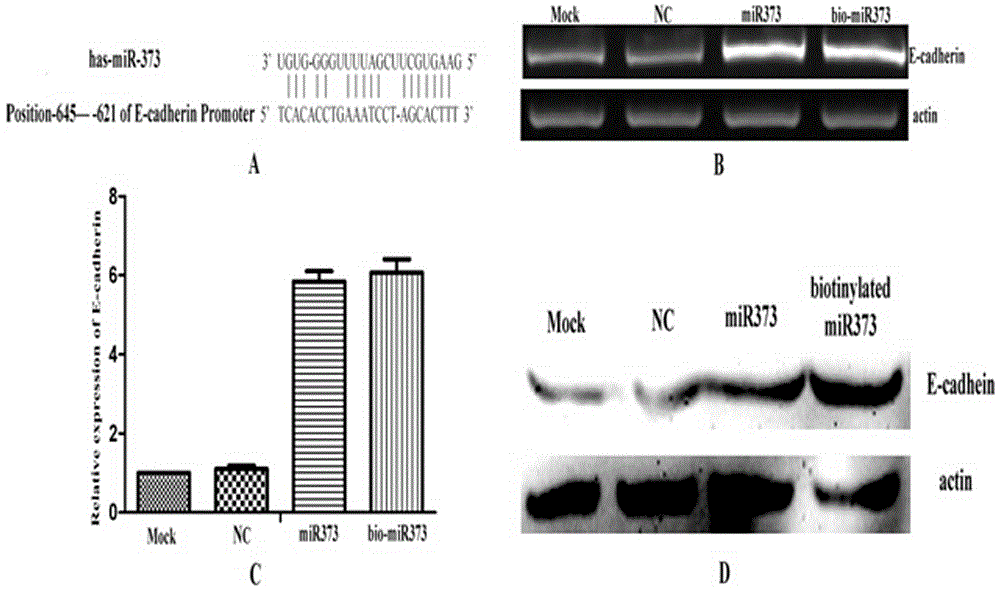
[0016] Example 1 miRNA biomarker and transfection
[0017] The 3' end of the antisense strand of miRNA-373 was biotin-labeled, transfected into cells using the transfection reagent Lipofectamine 2000, and the total RNA was extracted using Trizol 24 hours later. After reverse transcription into cDNA, the real-time quantitative PCR method was used to detect the regulatory effect of miRNA-373 on the expression of the target gene E-cadherin, and the amplification primers used were:
[0018] E-cadherin RTF: 5'-CCTGGGACTCCACCTACAGA-3',
[0019] E-cadherin RTR: 5'-GGATGACACAGCGTGAGAGA-3'.
[0020] actinRTF: 5'-CGTGGACATCCGCAAAGAC-3',
[0021] actinRTR: 5'-TCGTCATACTCCTGCTTGCTG-3'.
[0022] Biotinylated miRNA-373 sequence:
[0023] miRNA-373anti-3biotin: 5'-GAAGUGCUUCGAUUUUGGGGUGU-3'-biotin.
Embodiment 2
[0024] Example 2 Cell culture and cell transfection
[0025] Normal breast cancer cell MCF-7 was from ATCC Company, USA. Culture in DMEM complete medium (plus 10% newborn bovine serum, 2 mM L-glutamic acid, penicillin and streptomycin) in a 37°C, 5% CO2, 90% relative humidity incubator until the cell density reaches 50 %-60%. Biotin-labeled miRNA-373 was transfected into MCF-7 cells with Lipofectamine 2000 transfection reagent. Before transfection, the medium was replaced with serum-free and antibiotic-free medium, and after 6 hours of transfection, it was changed into medium with serum and antibiotics to continue culturing.
Embodiment 3
[0026] Example 3 Streptavidin-Biotin pull-down experiment
[0027] Biotin-labeled miRNA-373 was transfected into MCF-7 cells with liposome Lipofectamine2000 as above, and 270 μL of 37% formalin solution (final concentration of 1 %) at room temperature for 10 min to fully cross-link proteins and RNA. Then add 1mL 10x Glycine (glycine) at room temperature for 5min to terminate the reaction. Aspirate the liquid in the culture dish, wash the cells twice with 10mL pre-cooled 1xPBS, scrape the cells with 1mL 1xPBS (2% Cocktail, RNase Inhibitor 100U / mL), harvest the cells at 800g, 3min in a 1.5mL RNase-Free centrifuge tube middle. Add 500 μL lysis buffer to each tube of cells, mix well, and let stand on ice for 30 minutes. Ultrasonic treatment was performed to break up the genomic DNA. Ultrasound working conditions: 200W, 5 seconds of ultrasound, 5 seconds of interval, 15 times of ultrasound. After standing on ice for 5 minutes, centrifuge at 14000g for 30s at 4°C. Transfer the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com