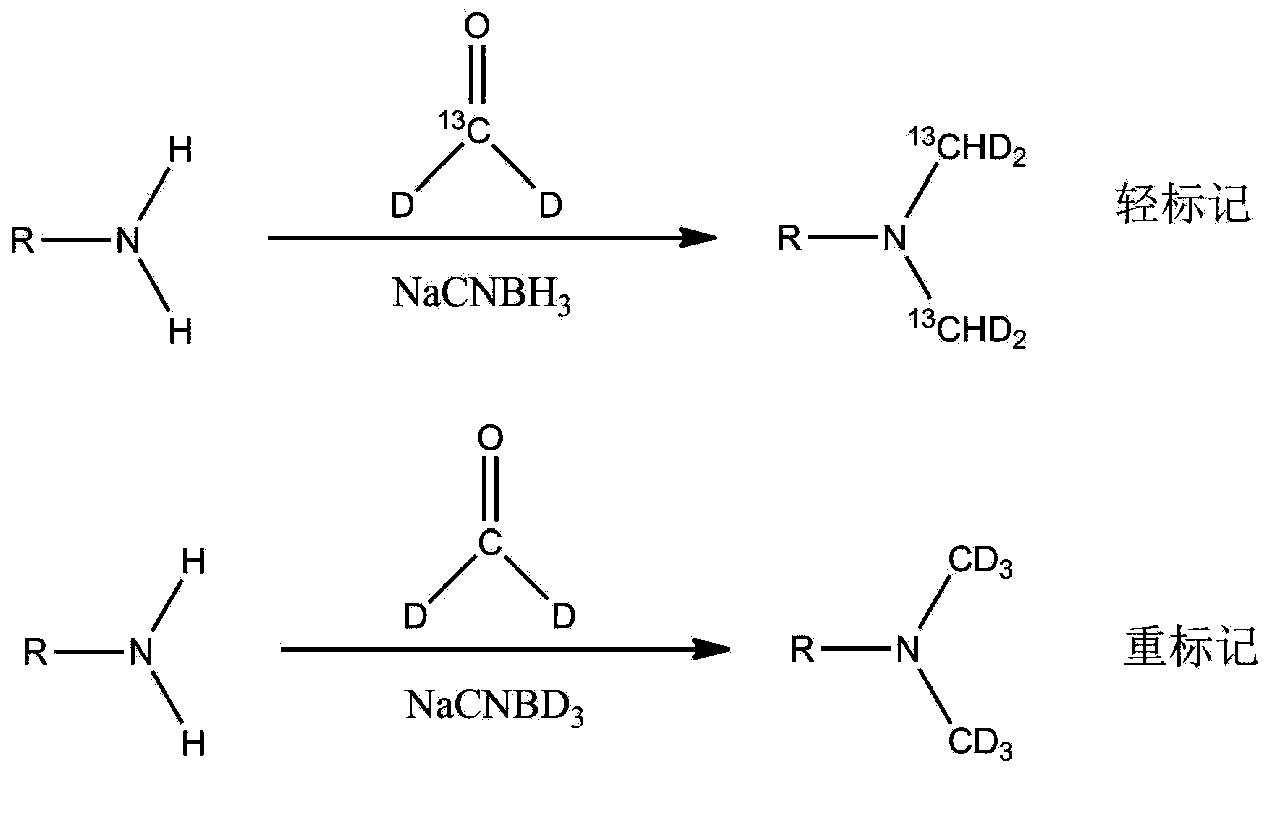
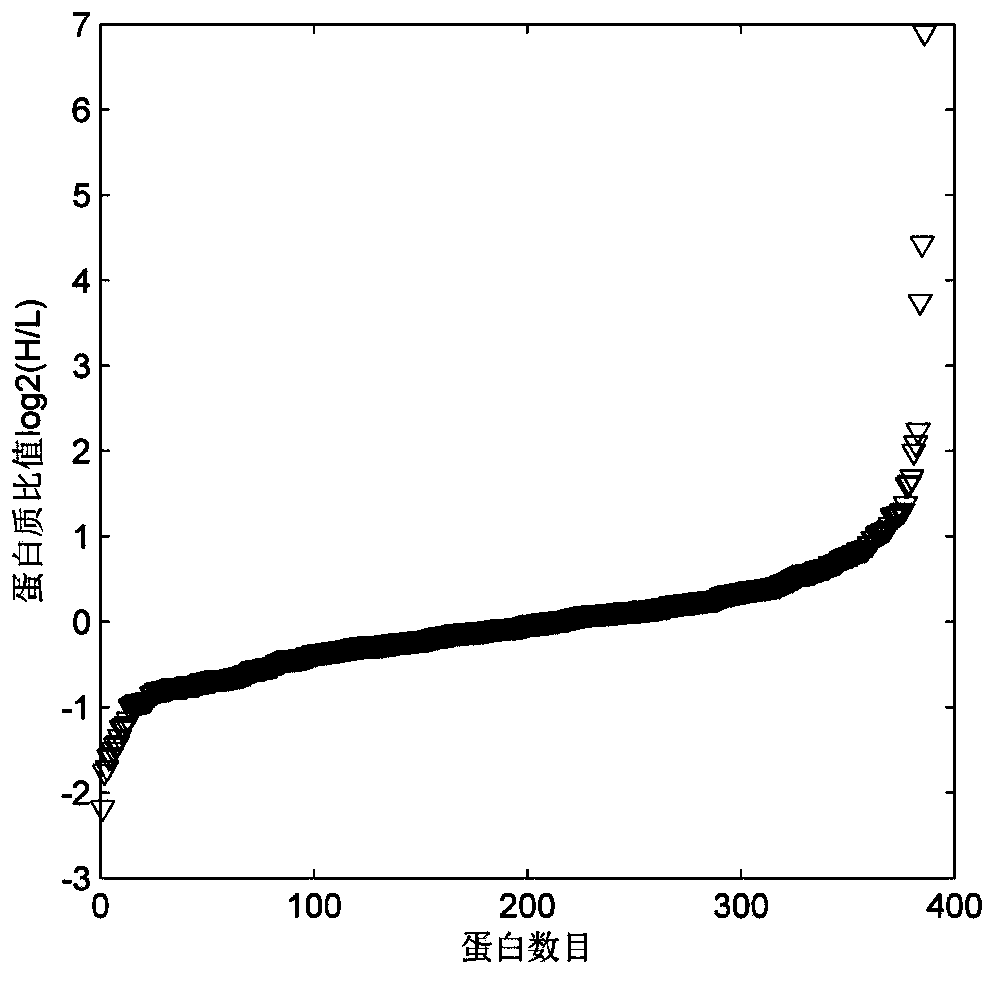
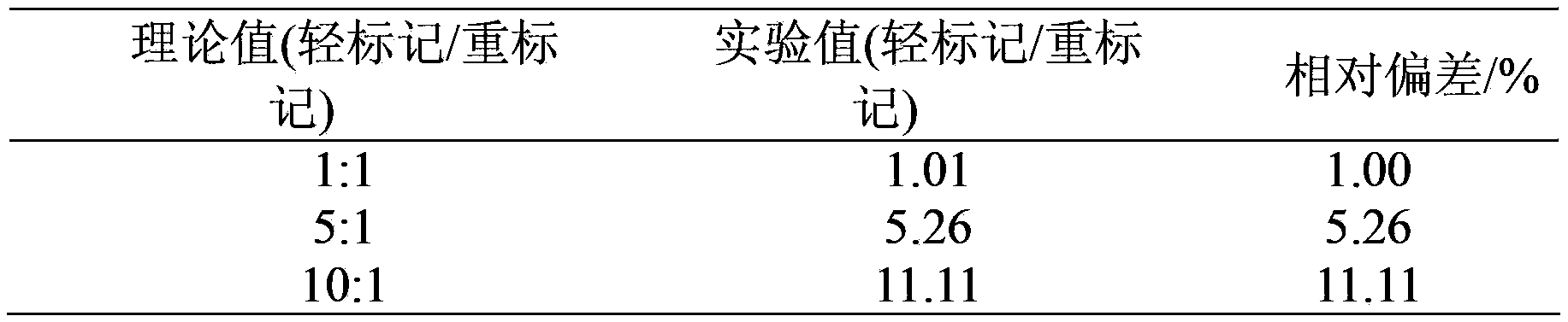
Protein quantitative method utilizing equiponderance dimethylation marking
A heavy dimethylation and protein technology, applied in the direction of measuring devices, instruments, scientific instruments, etc., can solve the problems of large sample loss, poor labeling efficiency, poor labeling efficiency and labeling selectivity, and achieve low labeling price, simple labeling, The effect of high labeling efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0022] 1. Protein extraction process: first wash the cells with 1×PBS until there is no blood. Use 2 mL of 8M urea plus commercial protease inhibitor cocktail (purchased from sigma) with a final concentration of 0.1% (v / v) as the lysate to suspend about 500 mg of mouse ascites-type liver cancer lymphatic high metastasis cell line (Hca -F) and about 500 mg of a low-metastatic cell line (Hca-P), the cell line was crushed with a tissue disruptor at a speed of 10,000 rpm. Then sonicate for 100 s. After ultrasonication, the protein extract was centrifuged at 25000g for 30min, precipitated with acetone at -80°C, discarded the supernatant by centrifugation, evaporated to dryness, reconstituted with 1mL 8M urea, and then determined the protein concentration by Bradford method. Store at -20°C until use.
[0023] 2. Protein sample processing process: 100 μL of 1 μg / μL Hca-P protein dissolved in 8M urea was added with dithiothreitol to a final concentration of 10 mM. After denaturation...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com