Medium and low temperature neutral tannase TanXZ7 and gene and application thereof
A tannase and neutral technology, applied in the field of genetic engineering, can solve problems such as slow progress, and achieve the effect of low acid-base low temperature adaptability and high activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Cloning of embodiment 1 Thielavia subthermophila XZ7 tannase coding gene tanXZ7
[0049] The strain Thielavia subthermophila XZ7 was collected from the root sand of vegetation in the desert area of Ningxia Autonomous Region. It can grow normally at 45°C, but cannot grow at 20°C. It is a typical thermophilic fungus.
[0050] Extraction of Thielavia subthermophila XZ7 genomic DNA:
[0051]Filter the mycelium cultured in liquid for 3 days into a mortar with sterile filter paper, add liquid nitrogen to make it pre-cooled, put the mycelia in the mortar and grind it into a white powder, transfer the white powder to a 50mL centrifuge Add 22.5mL of CTAB solution preheated at 65°C (add 2% mercaptoethanol before use) and 2.5mL of 20% SDS solution to the tube, lyse in a water bath at 65°C for 2 hours, invert and mix once every 10min, and centrifuge at 12,000rpm at 4°C 15min. The supernatant was extracted in chloroform / isoamyl alcohol (24:1) to remove impurities, and then 0.6 t...
Embodiment 2
[0060] The preparation of embodiment 2 recombinant tannase
[0061] The expression vector pPIC9 was subjected to double digestion (SpeI and NotI), and the gene tanXZ7 encoding tannase was double-digested (SpeI and NotI) at the same time, and the gene fragment encoding tannase was cut out and connected to the expression vector pPIC9 to obtain a shuttle containing The recombinant plasmid pPIC-tanXZ7 of Thielavia subthermophila XZ7 tannase gene tanXZ7 was transformed into Pichia pastoris GS115 to obtain recombinant Pichia pastoris strain GS115 / tanXZ7.
[0062] The GS115 strain containing the recombinant plasmid was inoculated in 400mL of BMGY culture medium, shaken at 250rpm at 30°C for 48h, and then collected by centrifugation. Then resuspend in 200mL BMMY medium, shake culture at 250rpm at 30°C. After 72 hours of induction, the supernatant was collected by centrifugation. Determination of tannase activity. The expression level of recombinant tannase was 10.52U / mL. SDS-PAGE ...
Embodiment 3
[0064] The activity analysis of embodiment 3 recombinant tannase
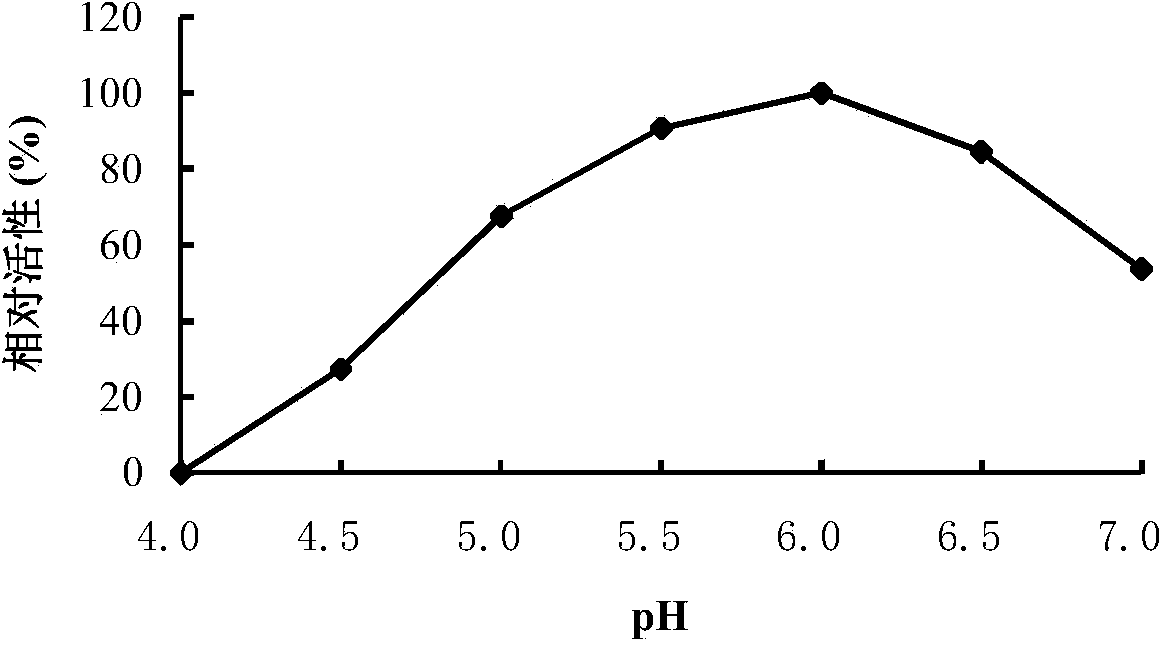
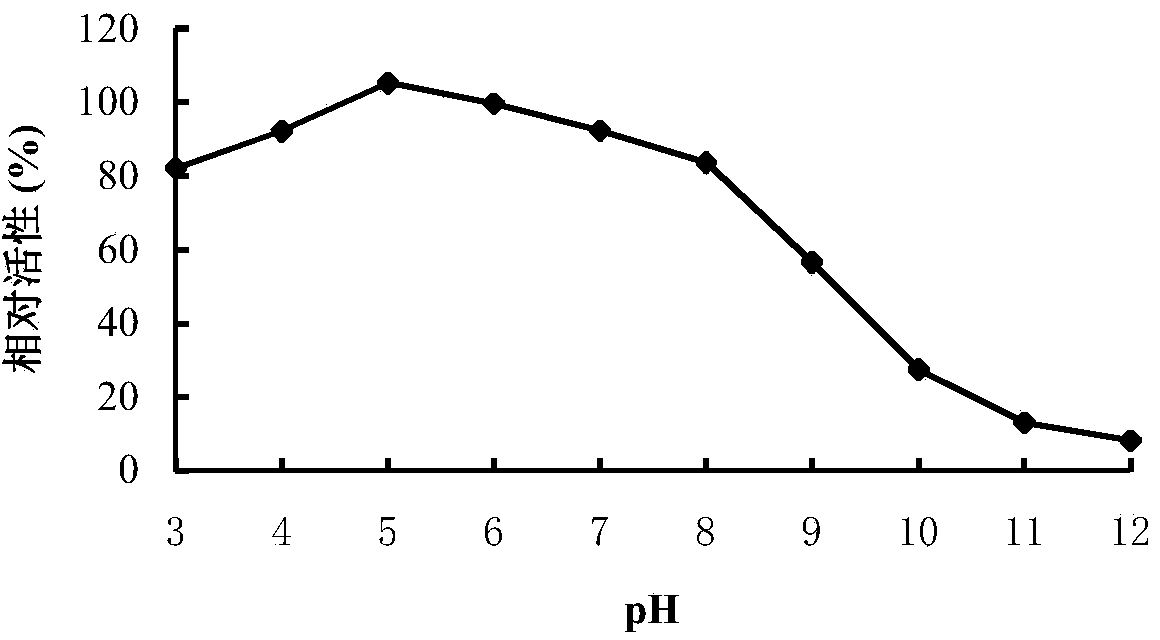
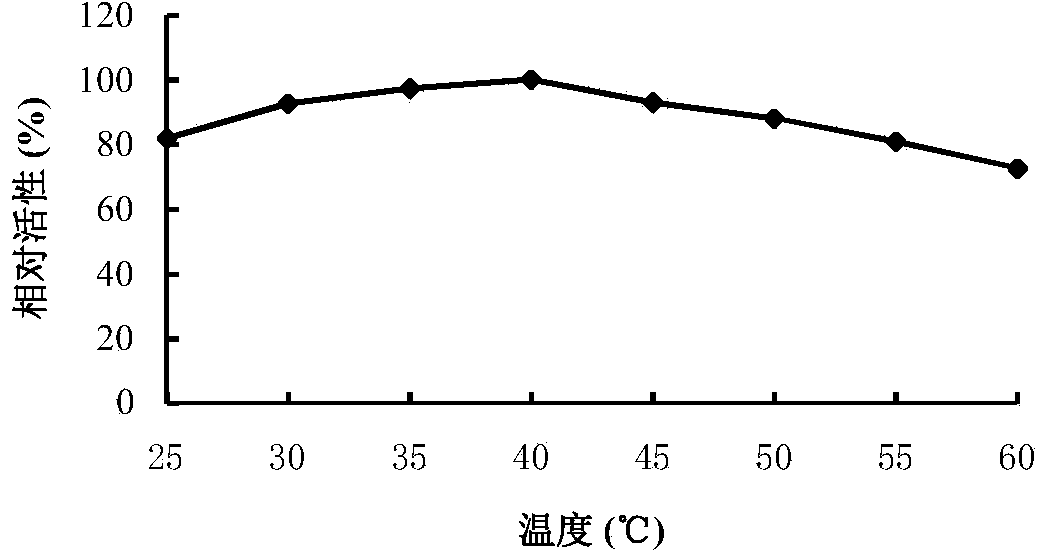
[0065] Rotanin Chromogenic Method: The specific method is as follows: at pH 5.0 and 40°C, a 500 μL reaction system includes 100 μL of an appropriate diluted enzyme solution, 400 μL of propyl gallate solution (1.25 mmol / L, pH 5.0), After reacting for 10 minutes, 300 μL of methanolic rhodanine solution (50 mmol / L) was added to terminate the reaction, and 200 μL of KOH (0.5 M) was added for color development after 5 minutes. After cooling to room temperature, the OD value was measured at 520 nm. One enzyme activity unit (U) is defined as the amount of enzyme that releases 1 μmol of gallic acid per minute under given conditions.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com