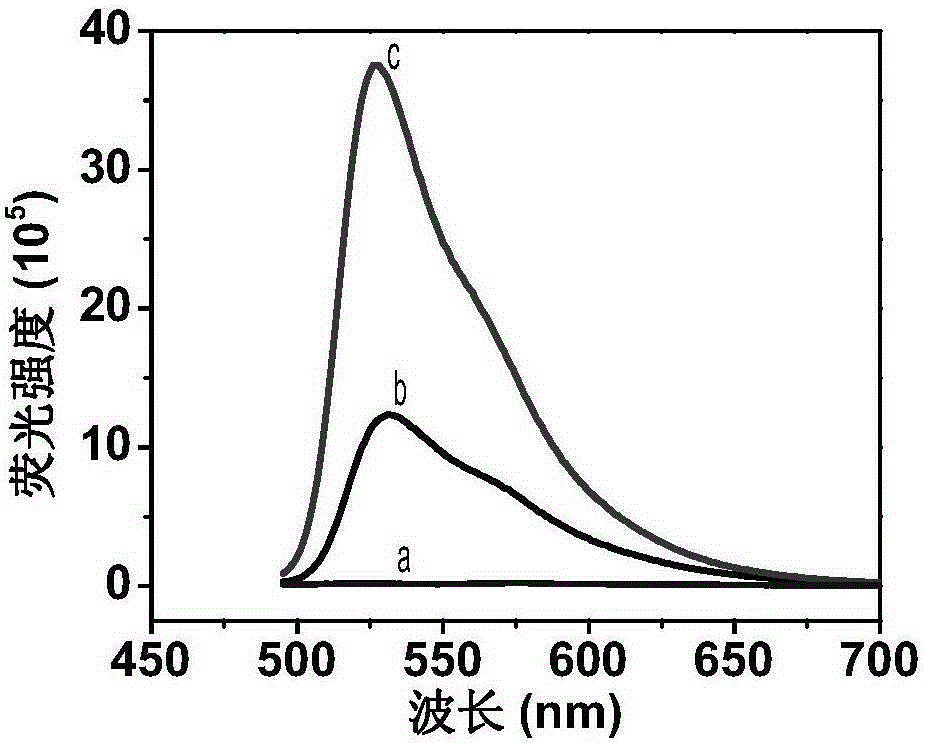
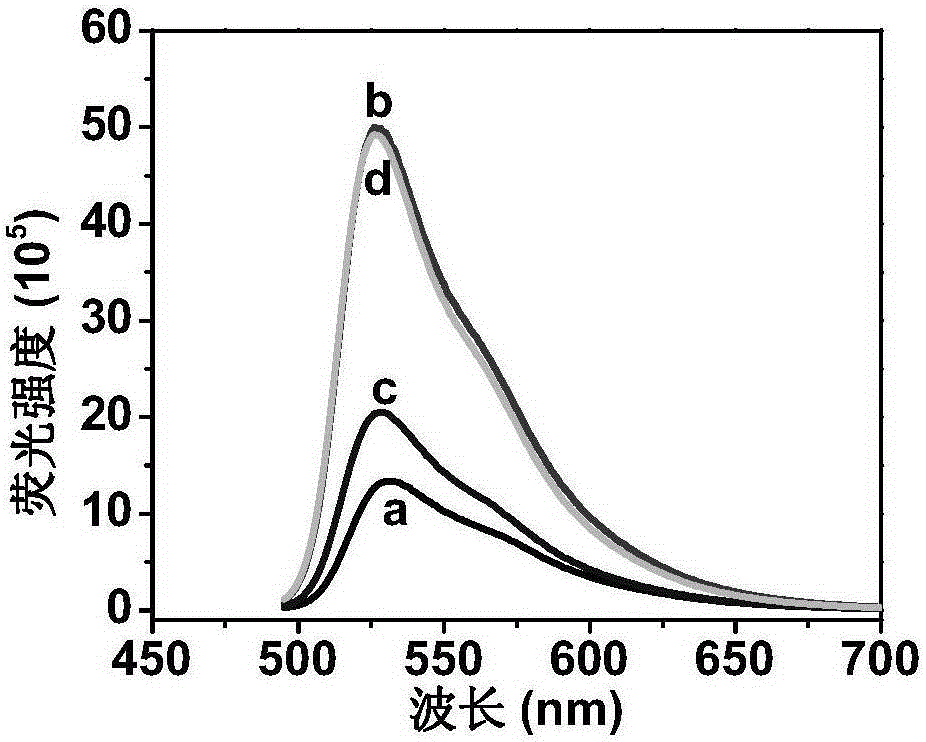
Label-free fluorescent detection of DNA methylation and methyltransferase activity by restriction endonuclease and exonuclease III
A technology of restriction endonuclease and exonuclease, which is applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem of not proposing to confirm methyltransferase at the same time, and avoid high detection costs. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] An analysis method based on restriction endonuclease and exonuclease III label-free fluorescent detection of DNA methylation, the detection steps are:
[0040] Hybridization step: in 50mM Tris-HCl (100mMNaCl, 5mMMgCl 2 , 0.1mg / mLBSA, 1mMDTTpH7.9) into the reaction system, add 5μL 10μM probe DNA and 5μL 10μM target DNA, mix thoroughly, react in a constant temperature water bath at 70°C for 10 minutes, cool down to room temperature naturally, and then add TO, The final concentration of thiazole orange in the buffer solution was 3.0 μM. After reacting at room temperature for one hour, the product solution was tested for fluorescence, and this solution was used as a control solution.
[0041] Methylation detection steps: add 7U methyltransferase and 50μMSAM to the DNA hybridization solution and mix well, react in a constant temperature water bath at 37°C for 1 hour, then add 10UHpaⅡ and 25UExoⅢ, mix well, and continue to maintain a constant temperature at 37°C After 2.5 ho...
Embodiment 2
[0044] An analysis method based on restriction endonuclease and exonuclease III label-free fluorescent detection of DNA methylation, the detection steps are:
[0045] Hybridization step: in 10mM Tris-HCl (300mMNaCl, 2mMMgCl 2 , 0.05mg / mLBSA, 0.05mMDTTpH7) into the reaction system, add a final concentration of 20nM probe DNA and 20nM target DNA, mix thoroughly, react in a constant temperature water bath at 70°C for 10 minutes, cool down to room temperature naturally, and then add TO, the final concentration of thiazole orange in the buffer is 0.7 μM. After reacting at room temperature for one hour, the product solution is used to measure fluorescence, and this solution is used as a control solution.
[0046] Methylation detection steps: add methyltransferase to the DNA hybridization solution with a final concentration of 1U / mL and 50μMSAM, mix thoroughly, and react in a constant temperature water bath at 37°C for 1 hour, then add HpaⅡ and ExoⅢ, the final concentration of HpaⅡ ...
Embodiment 3
[0049] An analysis method based on restriction endonuclease and exonuclease III label-free fluorescent detection of DNA methylation, the detection steps are:
[0050] Hybridization step: in 80mM Tris-HCl (500mMNaCl, 10mMMgCl 2 , 5mg / mLBSA, 10mMDTTpH7.5) into the reaction system, add a final concentration of 50nM probe DNA and 50nM target DNA, mix thoroughly, react in a constant temperature water bath at 70°C for 10 minutes, cool down to room temperature naturally, and then add TO, the final concentration of thiazole orange in the buffer is 1.6 μM. After reacting at room temperature for one hour, the product solution is used to measure fluorescence, and this solution is used as a control solution.
[0051] Methylation detection step: add the methyltransferase in the DNA hybridization solution to a final concentration of 10U / mL and 50μMSAM in the buffer, mix thoroughly, and react in a constant temperature water bath at 37°C for 1 hour, then add HpaII and The final concentration...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com