Transgenic constructs and their application in the preparation of epididymal head gene conditional knockout mouse model
A technology of transgenic and constructs, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
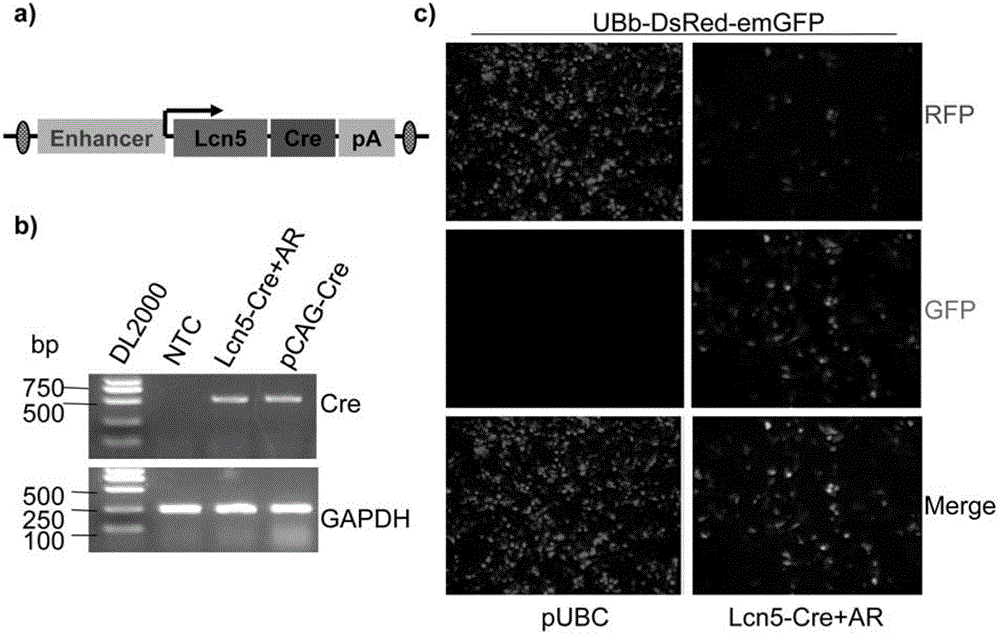
[0112] Construction of Lcn5(1.8)-Cre transgenic vector and in vitro activity verification
[0113] 1. The backbone vector used for the Cre recombinase transgene vector driven by a specific promoter in the head of the epididymis is a commercially available pUBC transgene vector
[0114] Firstly, the pUBC vector was transformed, and the 167bp UBC promoter sequence was cut with Mlu Ⅰ and Avr Ⅱ double restriction enzymes, and the Avr Ⅱ restriction site was replaced with Kpn Ⅰ.
[0115] Using the pBLCAT-5m-RABP plasmid (Lareyre, Thomas et al. 1999) as a template, through the primer pair: lcn5-pro-S: 5' ACGACGCGTCTACCTGGCCTCTCCTGCTCCT -3' (SEQ ID NO: 6) and lcn5-pro-A: 5' CGTGGTACCTGGGTTCAGCTCCCCACCAG 3' (SEQ ID NO: 7) for PCR reaction, the primer amplifies the 1.8 kb Lcn5(1.8)-Cre promoter sequence, the full length of the 1.8 kb sequence is shown in SEQ ID NO: 1. The PCR product and the pUBC transgene vector were digested with Mlu Ⅰ and Kpn Ⅰ restriction enzymes respectively, li...
Embodiment 2
[0121] Transgenic mouse construction and identification
[0122] 1. Use SgrD I and Fsp I restriction enzymes to linearize the Lcn5(1.8)-Cre transgenic vector prepared in Example 1, and prepare transgenic mice by prokaryotic microscopy. The background of the transgenic mice is C57BL / 6( Palmiter and Brinster 1985).
[0123] Rat tail DNA was extracted and passed through the primer pair
[0124] Tg-Cre-S:5' GCCTGCATTACCGGTCGATGC 3' (SEQ ID NO: 8) and
[0125] Tg-Cre-A:5' CAGGGTGTTATAAGCAATCCC 3' (SEQ ID NO: 9) was amplified by PCR to detect positive transgenic mice and establish a line.
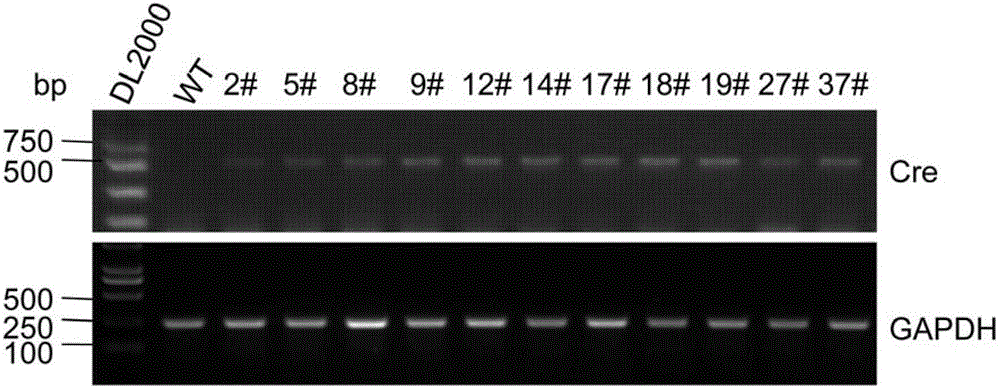
[0126] image 3 The identification and line establishment of the first transgenic mice were shown. A total of 38 first founder mice were born by pronuclear microinjection technique, among which 11 transgenic positive mice were identified by PCR technology, and WT was wild-type mice.
Embodiment 3
[0128] RT-PCR and quantitative PCR to detect the temporal and spatial expression characteristics of Cre recombinase mRNA in transgenic mice
[0129] Different tissues of Lcn5(1.8)-Cre transgenic mice were taken, and total RNA was extracted by referring to the operation method of TRIzol reagent (LifeTechnologies, USA), and the RNA was reversed by ReverTra Ace-α (FSK-100, TOYOBO) reverse transcription kit recorded as cDNA. The tissue-specific expression of Cre recombinase mRNA was detected by Tg-Cre-S and Tg-Cre-A (sequence as above).
[0130] In addition, the primer pair Lcn5-CDS-S: 5'TCAGCGAAGTACAAGGTCACC 3' (SEQ ID NO: 10) and Lcn5-CDS-A: 5'CATTGTTGTCCAAGCTCCG (SEQ ID NO: 11) were used to detect the expression of endogenous Lcn5 gene as a sample positive control.
[0131] Design the PCR primer pair cre-S: 5'GACCAGGTTCGTTCACTCATGGA 3' (SEQ ID NO: 12) and cre-A: 5'AACATTCTCCCCACCGTCAGTACG 3' (SEQ ID NO: 13), and detect Cre recombinase mRNA in the head of the epididymis by quan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com