A kit and method for detecting human parvovirus b19
A human parvovirus and parvovirus technology, applied in the biological field, can solve the problem of high detection cost, and achieve the effects of good repeatability, good application prospects and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] The design of embodiment 1 primer and the preparation of positive template
[0033] 1. Method
[0034] 1.1 Design and synthesis of primers
[0035] According to the sequence of the B19 virus on NCBI (NC_000883), primers were designed on the VP1 region of B19. The upstream primer (P1) was 5'-GGCACCTCTCAAAACACT-3' (SEQ ID NO: 1); the downstream primer (P2) was 5'-CCACTCCTTGCTGATACTC- 3' (SEQ ID NO: 2). Entrust Takara company to synthesize.
[0036] 1.2 Amplification of the B19 gene using the designed primers
[0037] 1.2.1 Nucleic acid extraction
[0038] The HighPureViralNucleicKit kit was used to extract B19-positive plasma and nucleic acid containing B19-positive plasma IVIG, and the nucleic acid of B19-negative plasma extracted by the kit was used as a negative control, and stored at -40°C.
[0039] 1.2.2 Amplification and identification
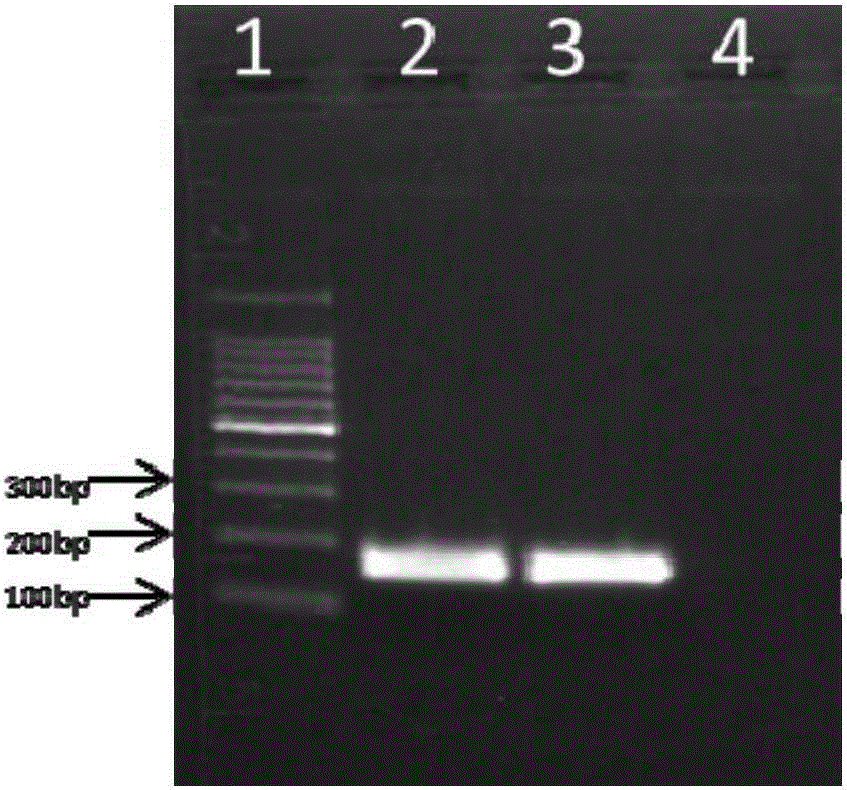
[0040] Using the genomic DNA of B19 as a template, using P1 and P2 as primers, PCR amplification:
[0041] Reaction system ...
Embodiment 2
[0054] The specific detection of embodiment 2 primers of the present invention
[0055] 1. Method
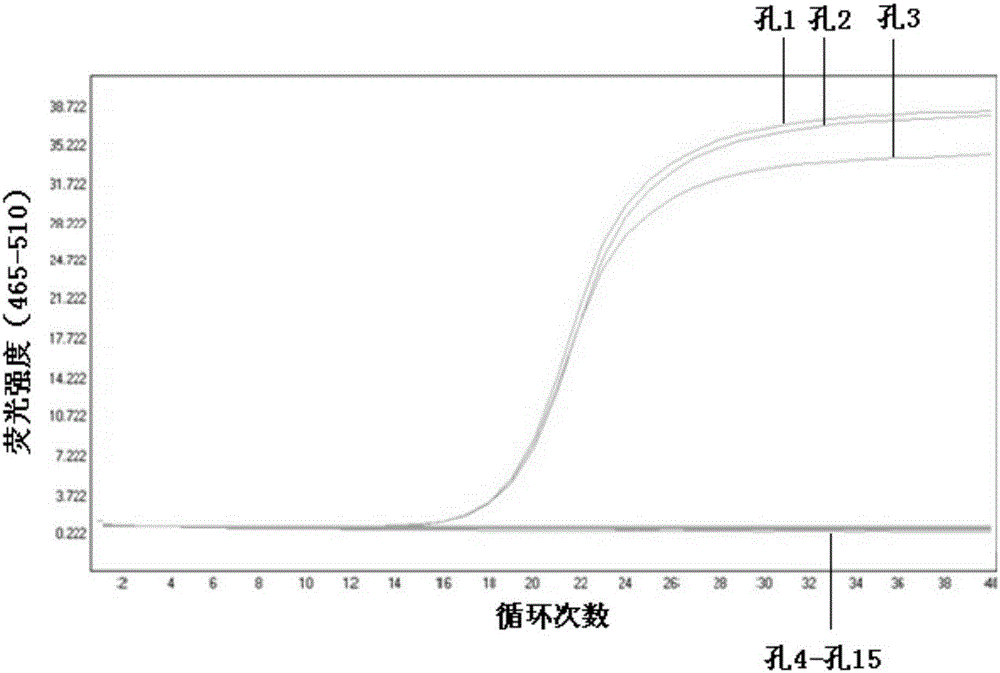
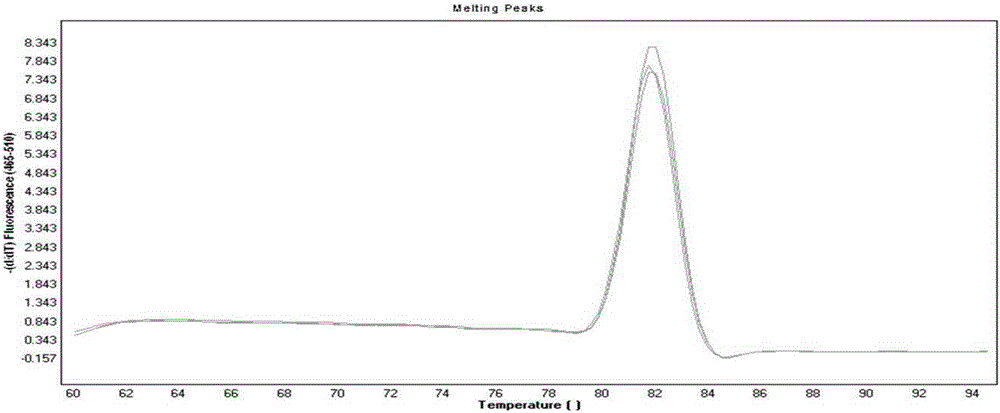
[0056] Extract the DNA of B19, PRV, BPV, MMV, and PK-15 as templates according to the method of 1.2.1 in Example 1, and perform PCR amplification. Three duplicate holes are set for each sample, and B19 (hole 1, hole 2, hole 3), PRV (hole 4, hole 5, hole 6), BPV (hole 7, hole 8, hole 9), MMV (hole 10, hole 11, hole 12), PK-15 (hole 13, hole 14, hole 15):
[0057] Reaction system (total volume 15 μl): EvaGreen master mix (SsoFast TM Supermix, 172-5201, Bio-rad company) 7.5 μl, upstream and downstream primers 0.5 μl, template DNA 1 μl (in ddH 2 O to make up 15 μl).
[0058] Reaction conditions: 95°C for 5min; 95°C for 10s, 52°C for 10s and 72°C for 10s, the last three steps were repeated for 40 cycles.
[0059] View the amplification curve.
[0060] 2. Results
[0061] like figure 2 As shown, fluorescence signals are collected at each cycle in the PCR process. As the num...
Embodiment 3
[0064] The sensitivity detection of embodiment 3 primers of the present invention
[0065] 1. Method
[0066] Dilute the B19 high-concentration positive plasma as shown in Table 1 to obtain samples containing different titers of B19;
[0067] According to the method of 1.2.1 in Example 1, sample nucleic acid was extracted respectively as a template, and PCR amplification was repeated 3 times at different times, and 8 parallel tubes were made at each concentration:
[0068] Reaction system (total volume: 15 μl): EvaGreen master mix 7.5 μl, upstream and downstream primers 0.5 μl, template DNA 1 μl (15 μl supplemented with ddH2O).
[0069] Reaction conditions: 95°C for 5min; 95°C for 10s, 52°C for 10s and 72°C for 10s, the last three steps were repeated for 40 cycles.
[0070] Check the amplification curve, if there is an amplification reaction curve within 40 CP values, it is counted as detected, and if there is no amplification reaction curve within 40 CP values, it is counte...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com