Genetically engineered bacterium capable of increasing yield of spinosads as well as construction method and application
A technology of genetically engineered bacteria and spinosyn, which is applied in the field of genetically engineered bacteria with high spinosyn production and its construction, and can solve problems such as unbalanced spinosyn synthesis pathways and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] Embodiment 1, gene element cloning and plasmid construction containing corresponding gene element
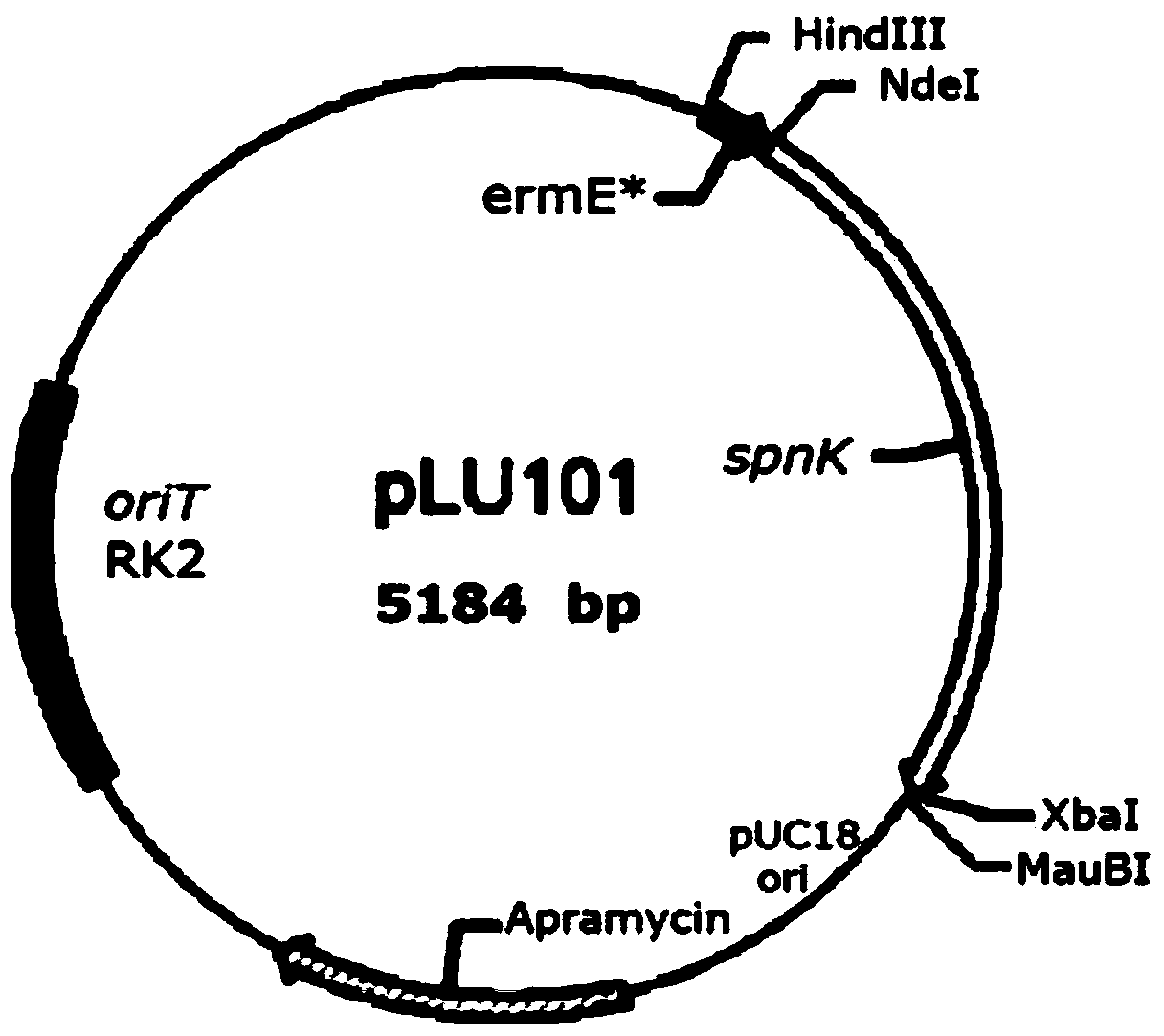
[0085] Digest the pIB139 plasmid containing the ermE* promoter (SEQ ID NO.23) with HindIII and XbaI. The enzyme used is Fermentas Fast Taq restriction endonuclease. The enzyme digestion system is as follows: 30ng of the target DNA fragment, add 2μL of 10×buffer , 1 μL each of restriction enzymes HindIII and XbaI (endonucleases are added at the end), and supplemented with distilled water to 20 μL. The enzyme digestion system was placed at 37°C for 30 min to obtain a 0.3 kb HindIII / XbaI fragment containing the ermE* promoter. This HindIII / XbaI fragment was inserted into plasmid pOJ260, resulting in pOJ261 plasmid.
[0086] Using the primer pair spnK-F-Ndel (SEQ ID NO.1) and spnK-R-Xbal (SEQ ID NO.2) from the chromosome of Saccharopolyspora spinosa (Saccharopolyspora spinosa) ATCC49460 (hereinafter referred to as Saccharopolyspora spinosa ATCC49460) The spnK gene sequence ...
Embodiment 2
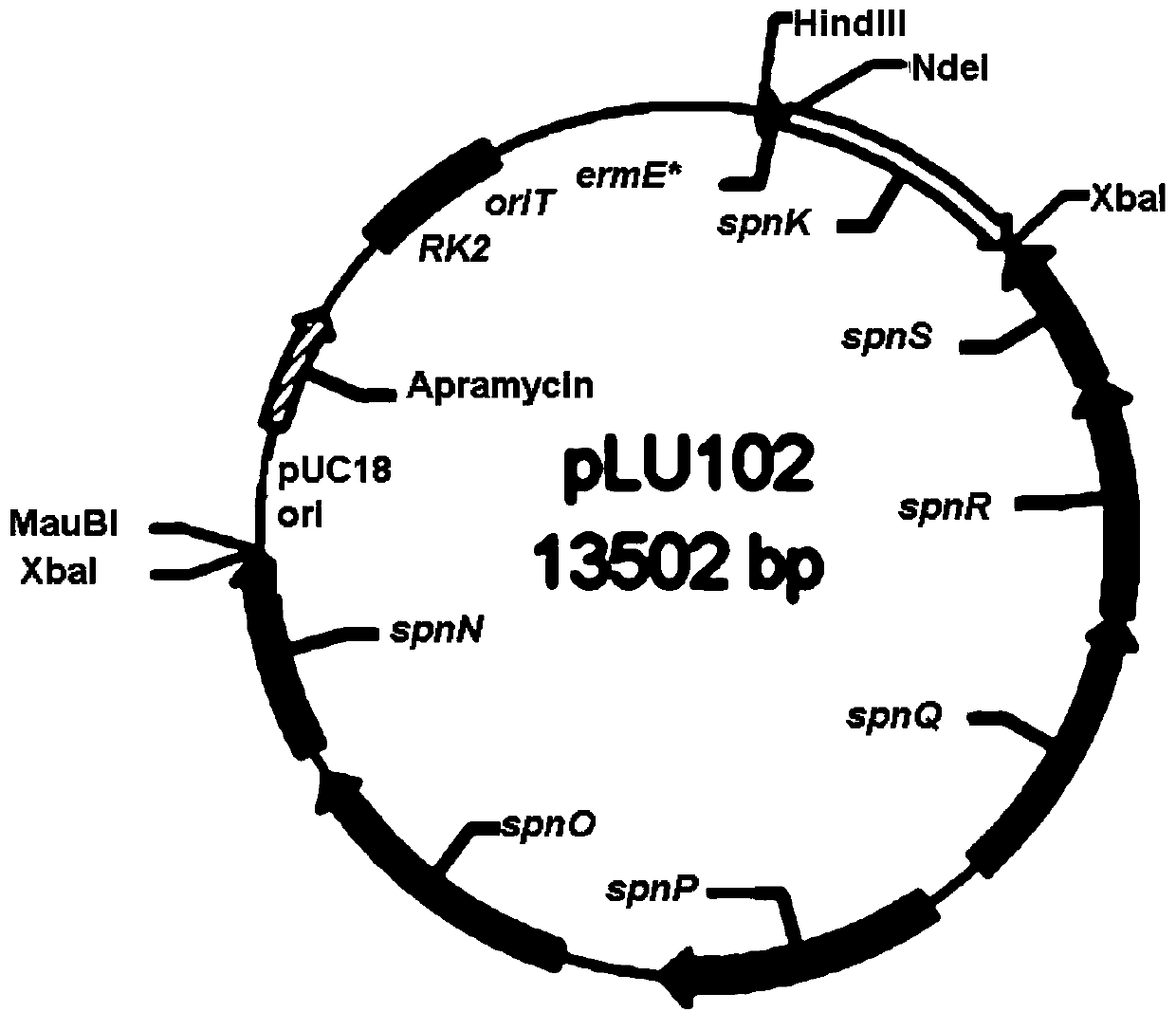
[0099] Embodiment 2, the construction of genetically engineered bacteria LU101, LU102 and LU104 that can improve spinosad output:
[0100]All constructed plasmids were introduced into S. spinosa from Escherichia coli S17-1 by combined transfer, and integrated into the chromosome by homologous recombination. The steps are as follows:
[0101] The plasmid pLU101 was used to transform competent Escherichia coli S17-1, and the obtained Escherichia coli was screened on an LB plate for apramycin resistance (ampramycin 100 μg / mL). The transformants were picked and cultured in 4 mL LB (ampramycin 100 μg / ml) with shaking at 37°C for 12 hours.
[0102] Pick an appropriate amount of Saccharopolyspora spinosa ATCC49460 from the slope and culture it in TSB liquid medium for about 72 hours at 30°C, transfer 1% of the inoculum to 50mL TSB and culture it for about 42 hours to make the bacterial liquid reach the late logarithmic growth period, centrifuge and pour off the supernatant , the cel...
Embodiment 3
[0129] Example 3, Fermentation Verification of Genetically Engineered Bacteria That Can Improve Spinosad Production
[0130] Strain medium:
[0131] Slant culture medium (g / L): casein peptone 2.5, glucose 5, beef extract 3, agar 20, magnesium sulfate 2, the balance is water, pH=7.5 before sterilization.
[0132] Seed medium (g / L): Glucose 10, N-Z-Amine A (acid hydrolyzed casein) 30, yeast extract 1.0, beef extract 1.0, MgSO42.0, the balance is water, pH=7.3 before sterilization.
[0133] Fermentation medium (g / L): glucose 68, milk peptone 25, cottonseed protein 22, corn steep liquor 14.5, methyl oleate 40, calcium carbonate 5, the balance is water, pH=7.5 before sterilization.
[0134] Shake flask fermentation culture: wash the mature spores with appropriate amount of sterile normal saline on the fresh plates of the genetically engineered bacteria LU101, LU102, LU104 that can increase the production of spinosad and the wild bacteria Saccharopolymona spinosa ATCC49460, and was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com