High-sensitivity method used for detecting and identifying human coronavirus
A technology of human coronavirus and kit, applied in biochemical equipment and methods, measurement/testing of microorganisms, resistance to vector-borne diseases, etc., can solve problems such as missed detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
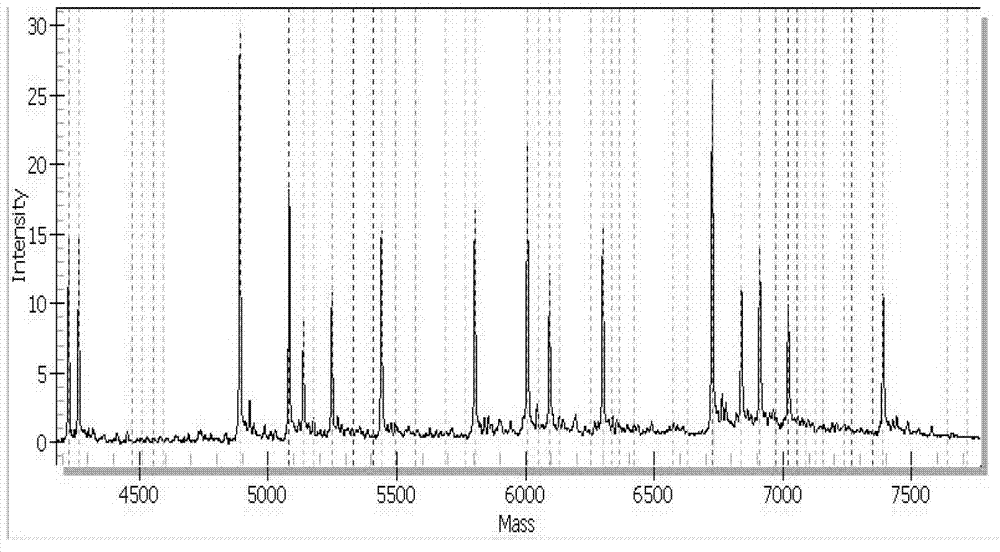
[0042] The present invention includes simultaneous detection of one or more of 6 known HCoVs, for HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS-CoV using 2-fold reaction for detection of specific sequence regions, for MERS-CoV was detected using a 4-plex reaction targeting specific sequence regions. The specific embodiments described here are only used to explain the present invention, not to limit the present invention.
[0043] According to the method for highly sensitive detection and / or identification of human coronaviruses provided by the embodiments of the present invention, six kinds of human coronaviruses to be detected include: HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS-CoV and MERS- CoV. Including the following steps:
[0044] (1) According to the whole genome sequence of the HCoV to be tested, select the specific conserved sequence of each HCoV, design PCR primers, and have a tag sequence of any combination of 10 bases (ACGTTGGATG) at the 5' end of the pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com