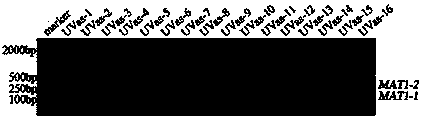Molecular detection method for rapidly differentiating mating type of Villosiclavavirens
A technology for the detection of rice smut bacteria and molecules, which is applied in the direction of microorganism-based methods, microorganism measurement/inspection, biochemical equipment and methods, etc., can solve the problems of cumbersome detection steps, achieve reduced detection steps, convenient and faster methods, The effect of saving testing cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0015] Clone acquisition mat1-2-1 Partial DNA sequence:
[0016] According to the Aspergillus oryzae bacteria amplified by Yokoyama et al. mat1-2-1 Gene 210bp DNA sequence (Genbank accession number: AB124632), design specific nested primers, they are:
[0017] SEQ ID NO.5:
[0018] M2W1: 5’-CAAAGAGGCGAATCCAGGA-3’
[0019] SEQ ID NO.6:
[0020] M2W2: 5’-CTGGGCCGAGCATGGAACCTTGA-3’
[0021] SEQ ID NO.7:
[0022] M2W3: 5’-ACAGGATCAAGCAGGCCCTCTT-3’
[0023] DNA Walking SpeedUp kit (Seegene) was used to amplify the 5’ end DNA sequence of the sequence and sequenced to obtain a total of 526 bp mat1-2-1 Partial DNA sequence of gene
[0024] SEQ ID NO.8:
[0025] mat1-2-1 Partial DNA sequence of gene
[0026] ccgctgtcct tgttagcttc gggatccatt ggacagggtg ccgtcgatgc ctggacgggc 60
[0027] agacagccgt gtggtggtgc tgcgtaccgt ttgcatcccc agcttggcgt gttttatttt 120
[0028] tgcgtctttc ttcggcttcc cggccgcctc agtttgcgtc gtgggtgctg ggcggactgg 180
[0029] ctgtgagggg gattccgctt tcgggggacg agatggggaa gtcgtcggcg gtggat...
Embodiment 2
[0036] Mating type gene mat1-1-1 Partial DNA sequence (Genbank accession number: JQ844754 ) Design primer pairs:
[0037] SEQ ID NO.1:
[0038] Vm1-F: 5’-GAAGTCGTATGCGTGCGAC-3’
[0039] SEQ ID NO.2:
[0040] Vm1-R: 5’-CTTGTTCCACAGGGTGGTCA-3’
[0041] The mating type gene of Aspergillus oryzae obtained according to Example 1 mat1-2-1 Partial DNA sequence (SEQ ID NO.8) design primer pair:
[0042] SEQ ID NO.3:
[0043] Vm2-F: 5’-CAATCTGCGCTTGGGTGTTC-3’
[0044] SEQ ID NO.4:
[0045] Vm2-R: 5’- GGAGCGACATAATACCGTCA -3’.
Embodiment 3
[0047] Using multiplex PCR detection method to determine the mating type of Aspergillus oryzae:
[0048] Collect hyphae and tissue samples to be tested. Including conidia monospora strains, ascospore monospora strains or sclerotia collected in the field.
[0049] CTAB was used to extract the genomic DNA of the required test sample and analyzed by 0.7% agarose gel electrophoresis.
[0050] Using multiple PCR amplification system to detect the mating type of Aspergillus oryzae. The amplification primer pairs are: SEQ ID NO.1 and SEQ ID NO.1; SEQ ID NO.3 and SEQ ID NO.4.
[0051] The multiplex PCR amplification system is: 10×PCR Buffer(Mg 2+ Free), 5uL; MgCl 2 (25mM), 3uL; dNTP Mixture (2.5mM each), 4 uL; template DNA (1-5ng / uL), 1uL; primers Vm1-F, Vm1-R and Vm2-F and Vm2-R, 1 uL each ; Taq DNA polymerase, 0.25uL. The PCR amplification program is: 94°C, 5min; 35×(94°C, 30s; 65.4°C, 30s; 72°C, 30s); 72°C, 5min; 4°C storage.
[0052] Take 5uL PCR amplification product and perform 2% ag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com