Method for preparation of S-adenosylmethionine
A technology of adenosylmethionine and amino acids, applied in the field of biochemical industry, can solve problems such as the lack of identification of SAM transporters and the biological safety of Escherichia coli
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0097] Embodiment 1, gene cloning and plasmid transformation
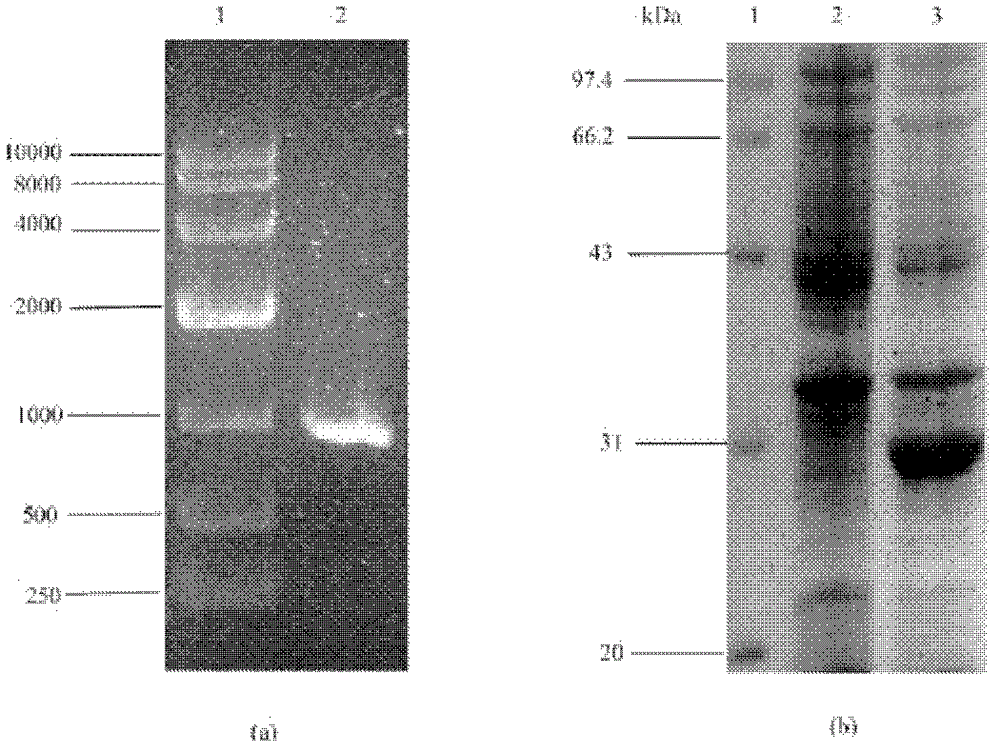
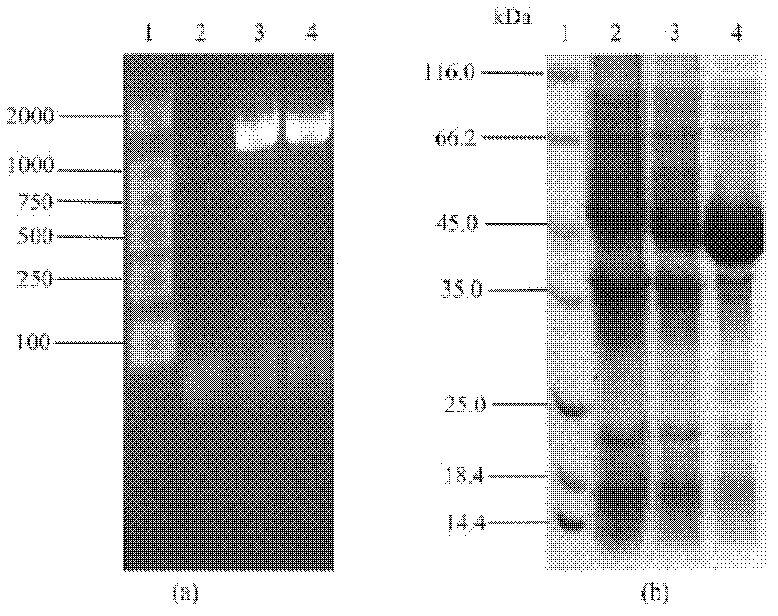
[0098] The principle of transforming the plasmid is: use the method of genetic engineering to insert pet8, metk and sam2 genes on pET21, and then clone the pet8 gene containing the regulatory sequence into p-metk and p-sam2 clones to realize the co-expression of transporter and synthetase Express. The schematic diagram of each transformed plasmid is shown in Figure 6 .
[0099] 1. Cloning, plasmid construction and expression of pet8 gene
[0100] Since there is no intron in the pet8 gene in the Saccharomyces cerevisiae genome, it can be directly amplified from the genome without RT-PCR. Primers were designed according to the gene sequence (SEQ ID NO: 1) of the pet8 gene in Gene ID: 855729, and the extracted Saccharomycs cerevisiae (Saccharomycs cerevisiae S288c) genomic DNA was used as a template for PCR amplification, and the upstream primer was pet8-1: 5' -CCC AAGCTT ATGAATAC TTTTTTTC-3' (SEQ ID NO: 7); do...
Embodiment 2
[0120] Example 2, the co-cloning expression of metk / sam2 and pet8
[0121] The three plasmids p-pet8, p-metk, and p-sam2 prepared above were further recombined and constructed, and the pet8 gene was placed on p-metk and p-sam2 to achieve co-expression.
[0122] The plasmid design primers of p-pet8 were carried out from the back of XbaI, so that sequences such as RBS were also placed in the multi-cloning site area. PCR amplification was performed using Saccharomyces cerevisiae S288c genomic DNA as a template, and the upstream primer was pt-pet-1:5'-CCC AAGCTT AATAATTTTGTTTAACT-3' (SEQ ID NO: 13); downstream primer is Pt-pet-2: 5'-TTT TTACGCTCTCAT TT-3' (SEQ ID NO: 14); wherein the single underline in the pt-pet-1 primer is the HindIII restriction site, and the double underline in the Pt-pet-2 primer is the XhoI restriction site, so that Connected to the carrier.
[0123] The amplification system is:
[0124]
[0125] The amplification program was: denaturation at 98°C ...
Embodiment 3
[0128] Extraction and purification of SAM in embodiment 3, fermentation product
[0129] Plate the identified E.coli Rosetta strains expressing metk, sam2, pet8, metk-pet8 and sam2-pet8 respectively, and inoculate a single colony in liquid LB (containing 100 μg / mL ampicillin), culture overnight, and 1 The ratio of % (v / v) was inoculated in liquid LB medium, cultivated at 280rpm and 37°C for 3 hours, and when the OD600 reached 0.6, IPTG was added to a final concentration of 0.1mmol / L. After induction for 3 hours, add L-methionine, so that the final concentration of methionine was 0.5g / L, and then samples were taken at 24h and 48h. Centrifuge the obtained bacterial liquid sample at 12000rpm for 10min, discard the precipitate, filter the supernatant with a 0.22um ultrafiltration membrane, and prepare for HPLC detection.
[0130] Referring to the method mentioned in Cai Heng et al. [HPLC—UV method for determination of SAM in fermentation broth; Cai Heng, Zheng Weigang, Wan Honggu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com