Method for preparation of S-adenosylmethionine
A technology for adenosylmethionine and strain production, which is applied in the field of preparing S-adenosylmethionine, and can solve the problems of lack of identification of SAM transporter, biosafety of Escherichia coli and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0097] Embodiment 1, gene cloning and plasmid transformation
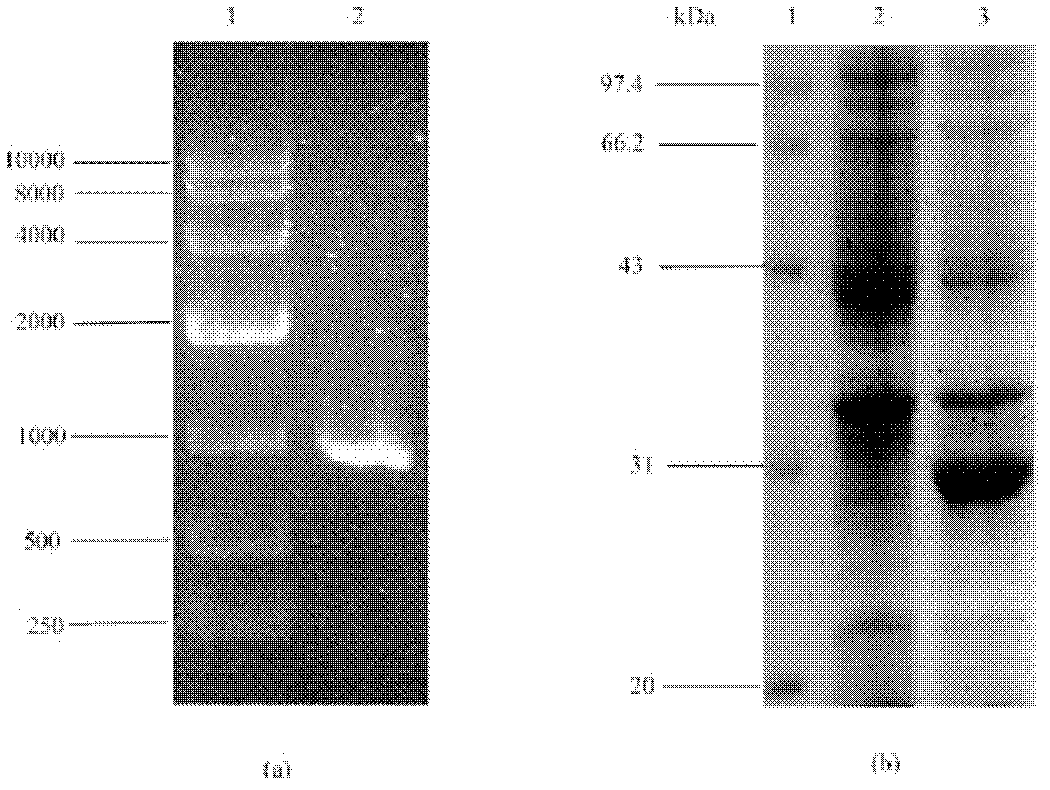
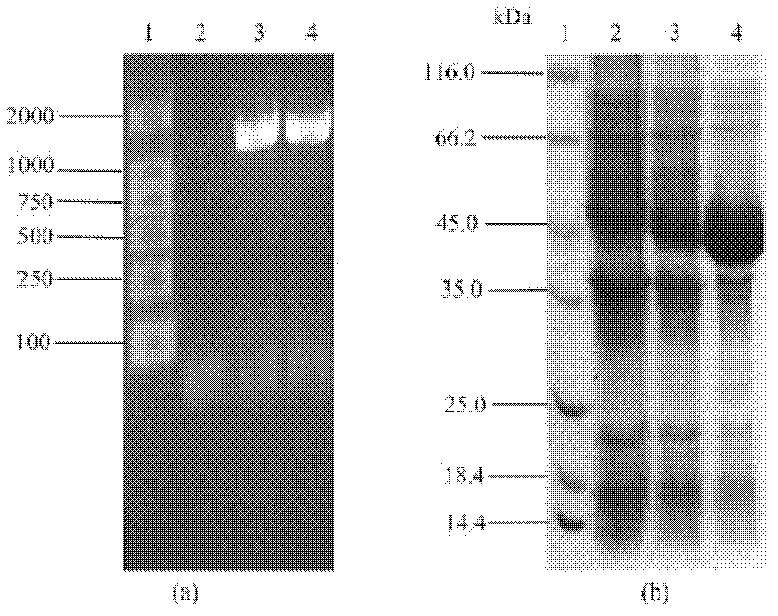
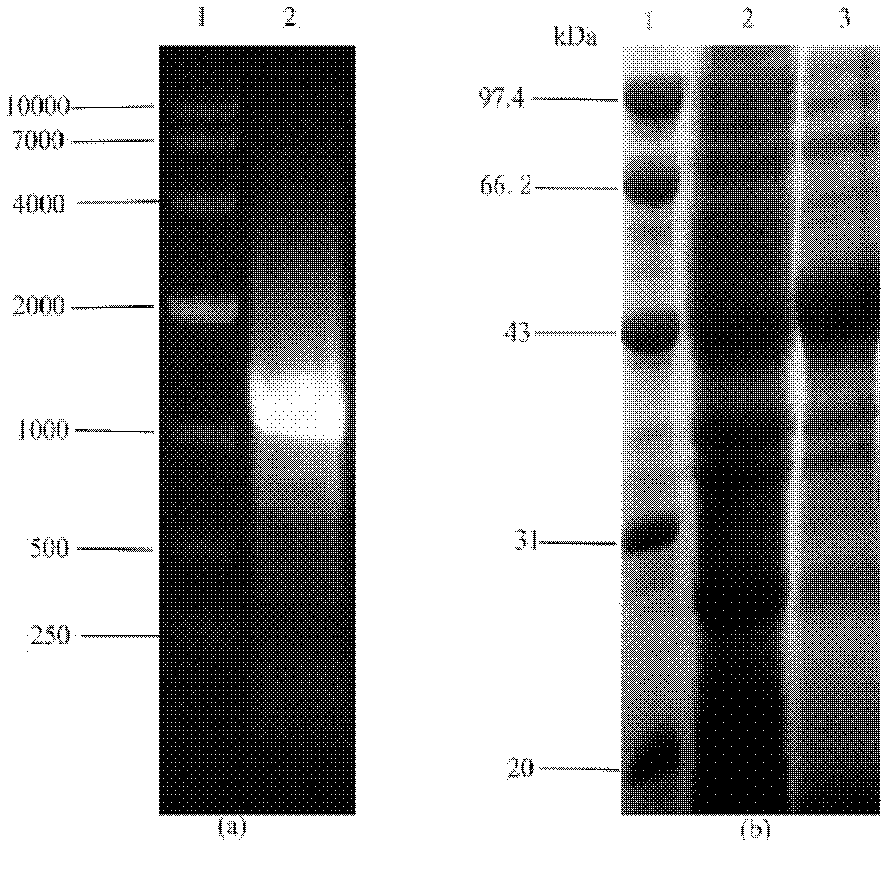
[0098] The principle of transforming the plasmid is: use the method of genetic engineering to insert pet8, metk and sam2 genes on pET21, and then clone the pet8 gene containing the regulatory sequence into p-metk and p-sam2 clones to realize the co-expression of transporter and synthetase Express. The schematic diagram of each transformed plasmid is shown in Figure 6 .
[0099] 1. Cloning, plasmid construction and expression of pet8 gene
[0100] Since there is no intron in the pet8 gene in the Saccharomyces cerevisiae genome, it can be directly amplified from the genome without RT-PCR. Primers were designed according to the gene sequence (SEQ ID NO: 1) of the pet8 gene in Gene ID: 855729, and the extracted Saccharomycs cerevisiae (Saccharomycs cerevisiae S288c) genomic DNA was used as a template for PCR amplification, and the upstream primer was pet8-1: 5' -CCC AAGCTT ATGAATAC TTTTTTTC-3' (SEQ ID NO: 7); do...
Embodiment 2
[0120] Example 2, the co-cloning expression of metk / sam2 and pet8
[0121] The three plasmids p-pet8, p-metk, and p-sam2 prepared above were further recombined and constructed, and the pet8 gene was placed on p-metk and p-sam2 to achieve co-expression.
[0122] The plasmid design primers of p-pet8 were carried out from the back of XbaI, so that sequences such as RBS were also placed in the multi-cloning site area. PCR amplification was performed using Saccharomyces cerevisiae S288c genomic DNA as a template, and the upstream primer was pt-pet-1:5'-CCC AAGCTT AATAATTTTGTTTAACT-3' (SEQ ID NO: 13); downstream primer is Pt-pet-2: 5'-TTT TTACGCTCTCAT TT-3' (SEQ ID NO: 14); wherein the single underline in the pt-pet-1 primer is the HindIII restriction site, and the double underline in the Pt-pet-2 primer is the XhoI restriction site, so that Connected to the carrier.
[0123] The amplification system is:
[0124]
[0125] The amplification program was: denaturation at 98°C ...
Embodiment 3
[0128] Extraction and purification of SAM in embodiment 3, fermentation product
[0129] Plate the identified E.coli Rosetta strains expressing metk, sam2, pet8, metk-pet8 and sam2-pet8 respectively, and inoculate a single colony in liquid LB (containing 100 μg / mL ampicillin), culture overnight, and 1 The ratio of % (v / v) was inoculated in liquid LB medium, cultivated at 280rpm and 37°C for 3 hours, and when the OD600 reached 0.6, IPTG was added to a final concentration of 0.1mmol / L. After induction for 3 hours, add L-methionine, so that the final concentration of methionine was 0.5g / L, and then samples were taken at 24h and 48h. Centrifuge the obtained bacterial liquid sample at 12000rpm for 10min, discard the precipitate, filter the supernatant with a 0.22um ultrafiltration membrane, and prepare for HPLC detection.
[0130] Referring to the method mentioned in Cai Heng et al. [HPLC—UV method for determination of SAM in fermentation broth; Cai Heng, Zheng Weigang, Wan Honggu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com