Method for rapidly detecting microsatellite markers of Charybdis feriatus
A technology of microsatellite markers and detection methods, which is applied to the determination/inspection of microorganisms, biochemical equipment and methods, etc., which can solve the problems of restricting the development of rust spots and no microsatellite markers of rust spots, and achieve clear amplification bands Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
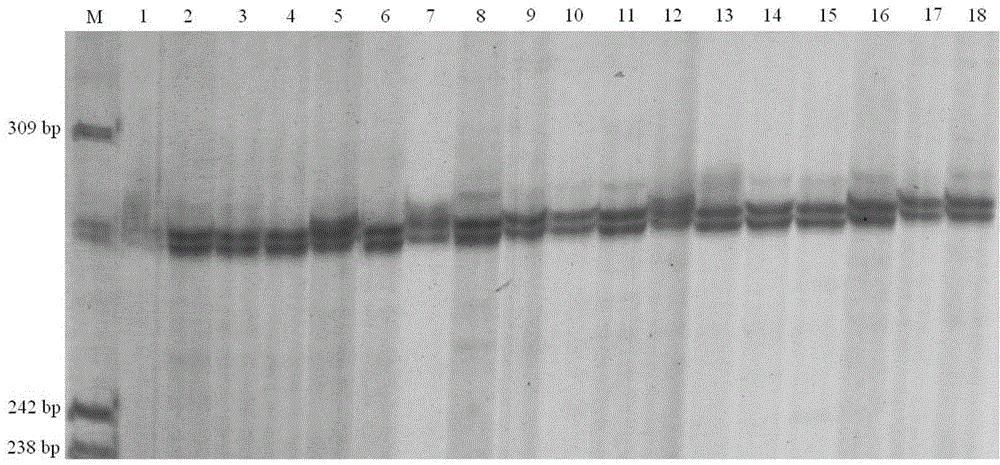
Image
Examples
Embodiment 1
[0034] 1. Extraction of Genomic DNA
[0035] Cut the muscle tissue (100-150mg) of Rust spotted worm and place it in 500μl tissue extract solution (10mmol / L Tris-Cl, pH8.0; 100mmol / L EDTA, pH8.0; 100mmol / L NaCl; 0.5%SDS) Cut into pieces, add proteinase K (final concentration 20mg / ml) and RNase A (final concentration 100μg / ml), mix thoroughly, digest at 55°C for 2-3 hours until clear; mix with phenol, chloroform and isoamyl alcohol solution (volume ratio 25:24:1) twice; then precipitate DNA with 2 volumes of absolute ethanol, wash with 70% ethanol and dry naturally, then dissolve in 50 μl TE buffer (10mmol / L Tris- HCl, pH8.0; 10mmol / LEDTA, pH8.0); finally the DNA concentration was diluted to 100ng / μl, and stored at -20°C for later use;
[0036] 2. Functional gene sequence search and microsatellite primer design
[0037] Search and download the functional gene sequence of Rust spotted cat from the GenBank database, and use the biological software SSRHUNTER1.3 to search for micr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com