Double-stranded nucleic acid and application thereof, and ribonuclease detection method
A ribonuclease, double-stranded nucleic acid technology, which is applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. Use value, wide applicability, simple operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] This example is used to illustrate the sequence structure of the oligoribonucleotide used in the present invention and the optical parameters of the fluorescent group. Entrust Guangzhou Ruibo Biotechnology Co., Ltd. to synthesize a series of oligoribonucleotides. A fluorophore is labeled at the 5' end of the oligoribonucleotide. Complementary oligoribonucleotides can be annealed to form double-stranded ribonucleic acid according to the method in "Molecular Cloning: A Laboratory Manual". Table 1 shows the sequences of the synthesized oligoribonucleotides. Wherein, in the oligoribonucleotide represented by SEQ ID NO: 11, the pentose sugar of the guanylic acid residue at position 3 is modified with 2'-oxymethyl (2'-OME). In the oligoribonucleotide represented by SEQ ID NO: 12, the pentose sugar of the uridine acid residue at position 14 is modified by 2'-fluoro substitution (2'-Fluro substitution). In the oligoribonucleotide represented by SEQ ID NO: 13, the pentose sug...
Embodiment 2
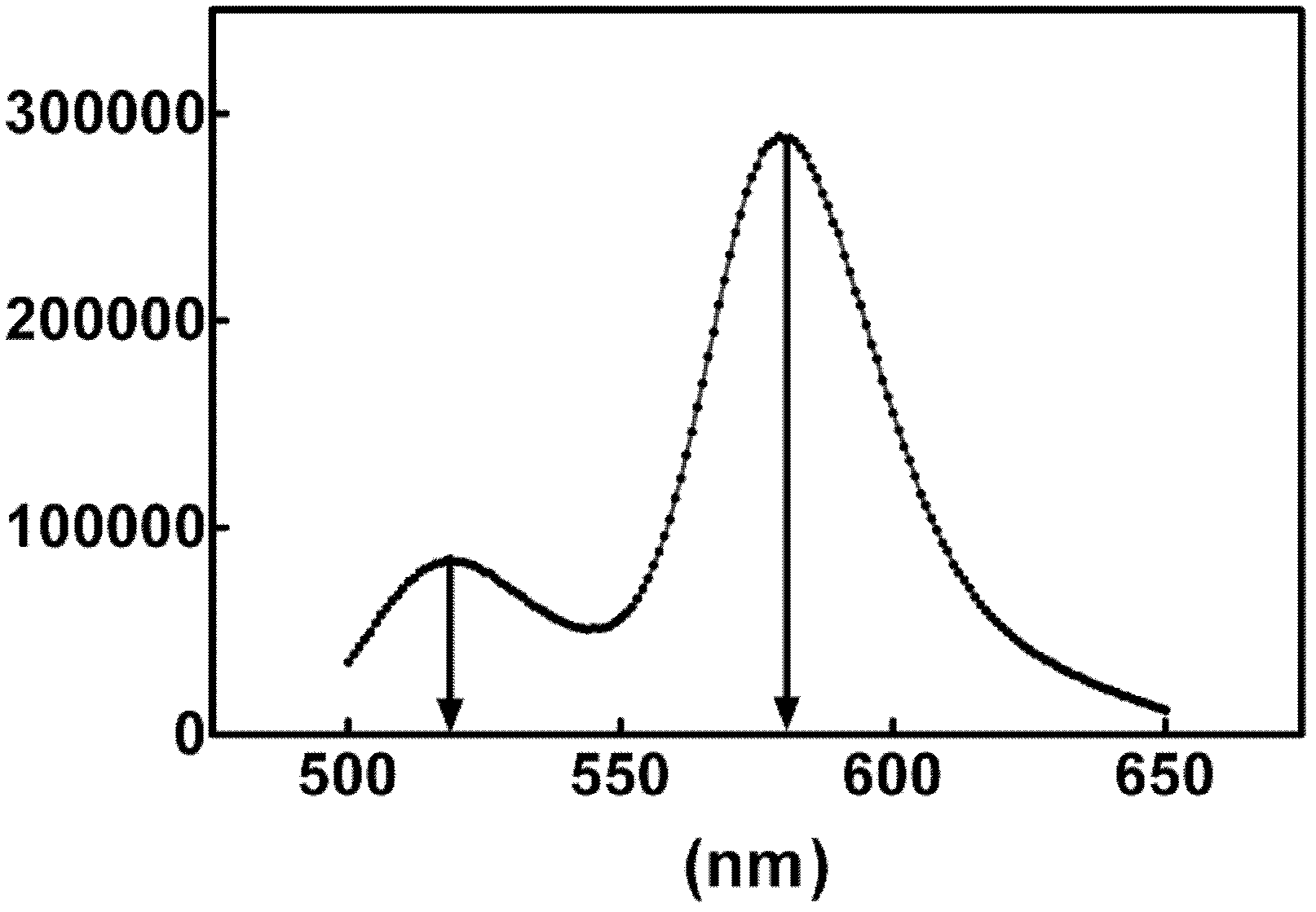
[0084] This example is used to illustrate the identification of the fluorescence resonance energy transfer phenomenon of the present invention. In the technical scheme for detecting ribonuclease levels by using fluorescence resonance energy transfer method provided by the present invention, a key condition is that labeling can occur between the fluorescent donor group and the fluorescent acceptor group at both ends of the double-stranded nucleic acid substrate. Fluorescence resonance energy transfer.
[0085] In order to verify this, at first the oligoribonucleotide (5'-FAM-AUGAGCCUGAUUU) of SEQ ID NO:1 and its complementary SEQ ID NO:2 oligoribonucleotide (UACUCGGACUAAA-TAMRA-5' ) anneal to form double-stranded ribonucleic acid substrate DS1. Take 4 μl of DS1 and add it to 2ml fluorescence resonance energy transfer buffer (0.01M Tris-HCl, pH 7.4, 0.002M MgCl 2 ), the final concentration of DS1 was 10nM. Subsequently, the reaction system is added into a quartz detection cup...
Embodiment 3
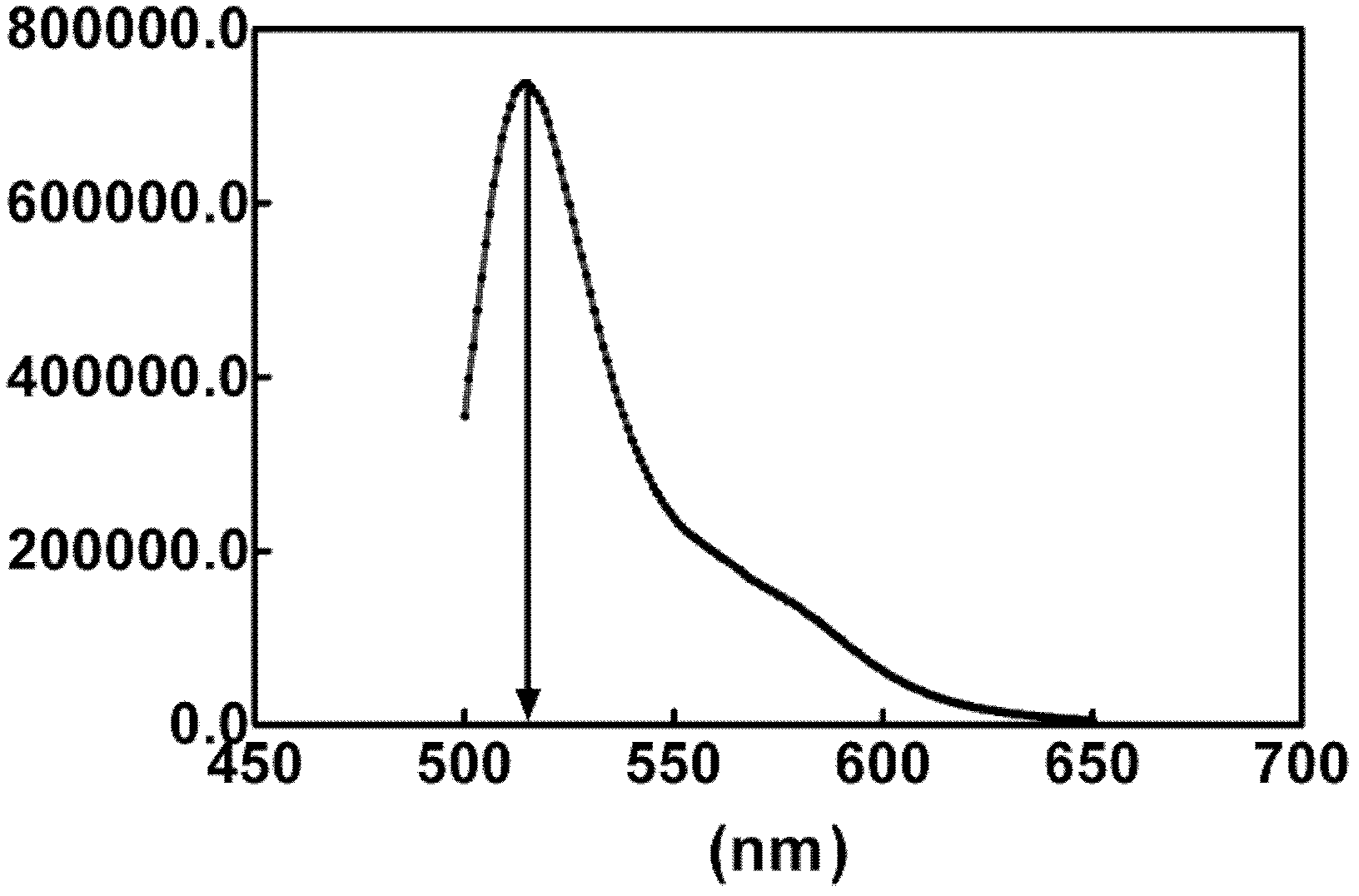
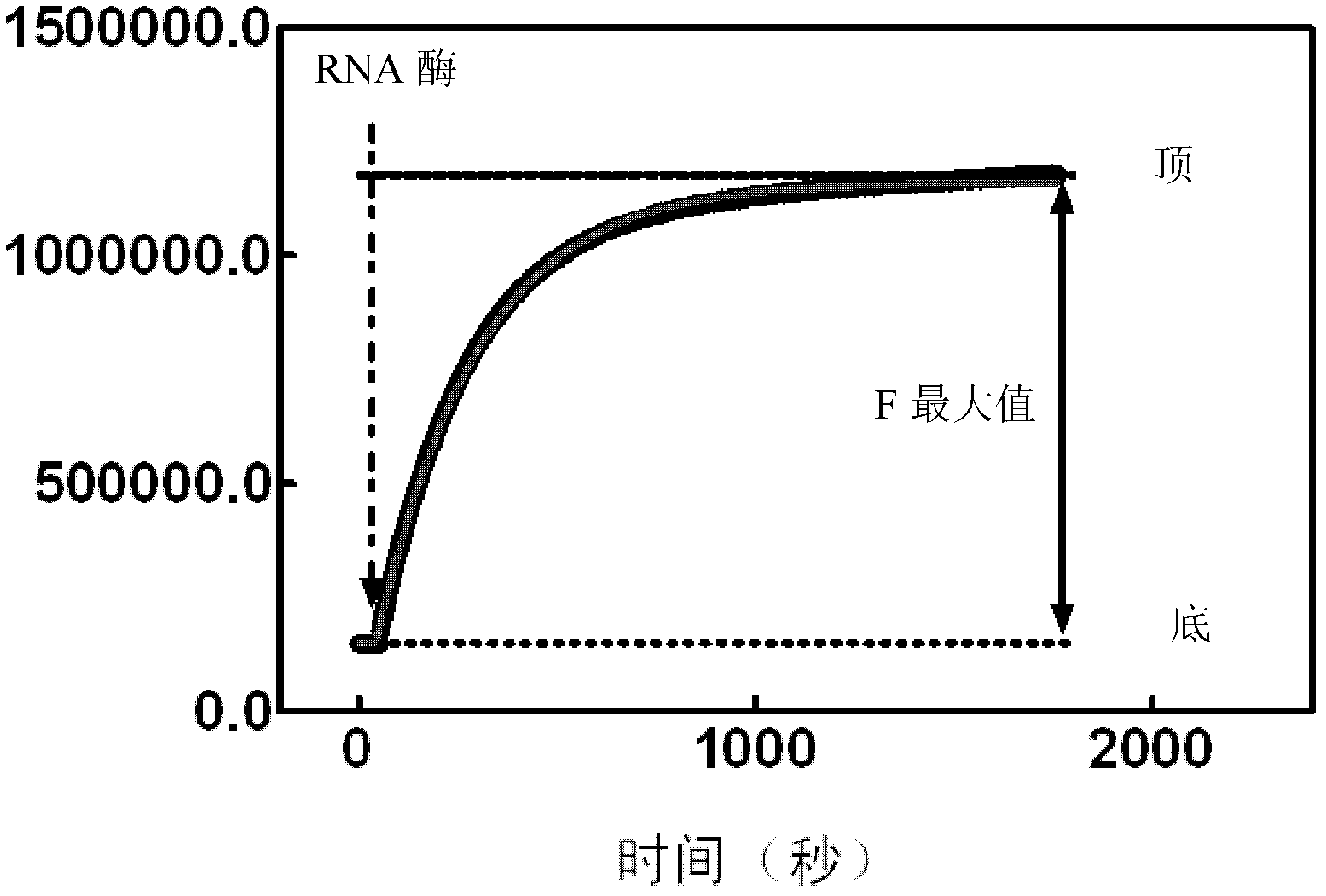
[0088]This example is used to illustrate the method for detecting ribonuclease by using the fluorescence resonance energy transfer method of the present invention. In order to utilize the fluorescence resonance energy transfer method to detect the activity and content of ribonuclease in the sample, take 4 μ l of double-stranded RNA substrate DS1 and join in 2 ml fluorescence resonance energy transfer buffer (0.01MTris-HCl, pH 7.4, 0.002M MgCl 2 ), the final concentration of DS1 was 10nM. Subsequently, the reaction system is added into a quartz detection cup of a fluorescence spectrometer, and a micro-magnetic rotor is placed in the quartz cup. After adding 20 μl of RNase A at a certain concentration to the reaction system, the reaction system was continuously excited at 480 nm and detected at 515 nm at the same time, and the fluorescence intensity values at intervals of seconds were obtained. Using these values to plot the curve of time versus fluorescence intensity, the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com