Laccase gene as well as encoded protein and application thereof
A technology of laccase and gene, applied in the field of laccase gene and its encoded protein and its application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Bacillus vallismortis fmb-103 cells (CGMCC No.6198) were collected by centrifugation, and the genomic DNA of Bacillus vallismortis fmb-103 was extracted with Shanghai Sangon Genomic DNA Extraction Kit.
[0032] Design two primers based on the registered Bacillus laccase gene (No.GU972592.1) in the Genebank database:
[0033] Upstream primer F-1: 5'-ATGACACTTGAAAAATTTGTGGATGC-3' (SEQ ID NO.3);
[0034] Downstream primer R-1: 5'-TTATTTATGGGGATCAGTTATATC-3' (SEQ ID NO.4);
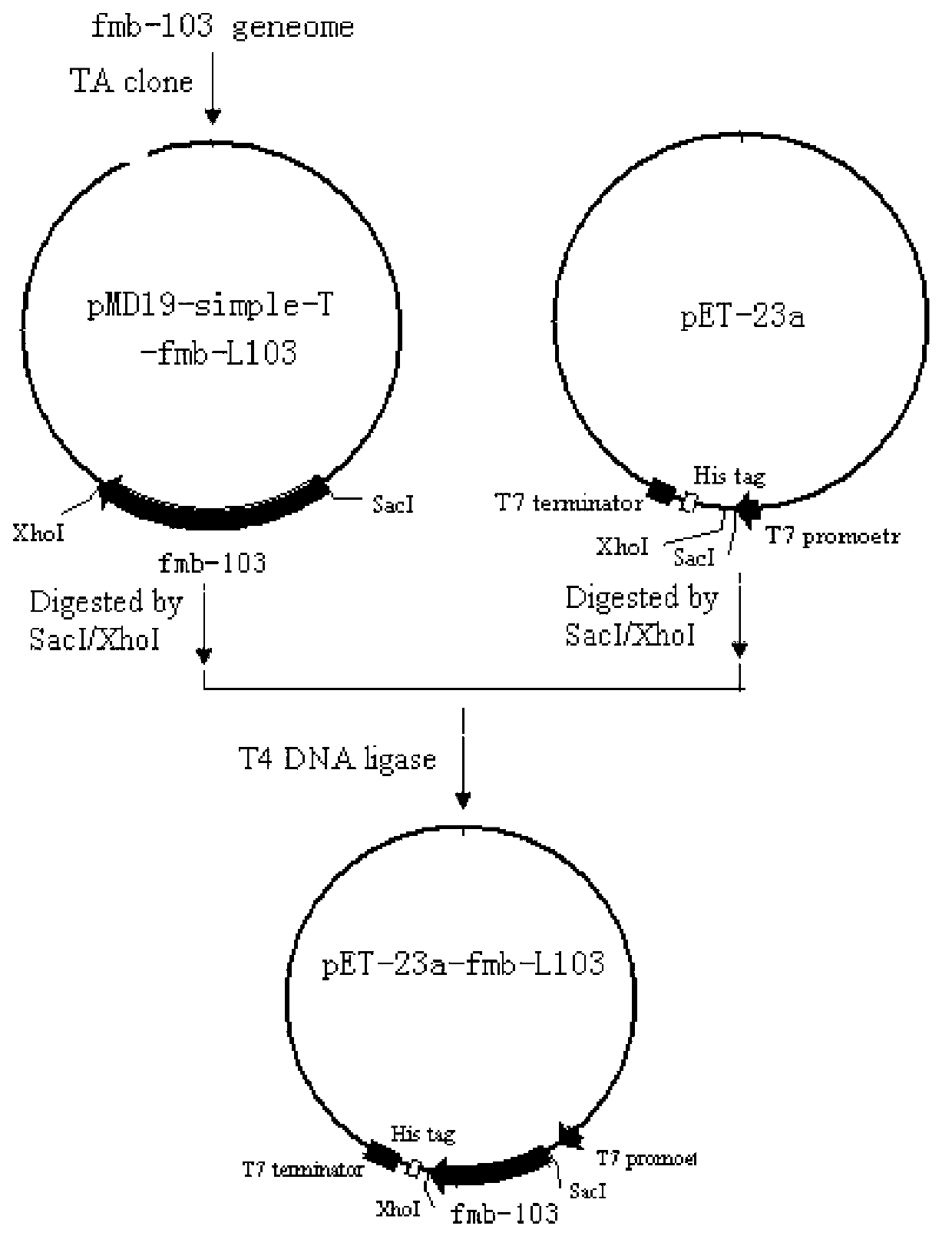
[0035] In a 50 μl system, the final concentration of each primer is 1 μM, the final concentration of dNTPs is 0.2 mM, fmb-103 strain genomic DNA 10 ng, 2 U Pfu DNA polymerase. The amplification program was 94°C for 3min; 30×(94°C for 40s, 53°C for 50s, 72°C for 90s); 72°C for 10min. Agarose gel electrophoresis, gel cutting, recovery by Shanghai Sangon kit, connection of the recovered PCR product with TaKaRapMD19-simple-T vector, transformation of E.coli DH5α, and spreading on LB containing IPTG, X-gal...
Embodiment 2
[0037] According to the obtained laccase gene sequence, two primers were designed, the upstream primer plus the SacI recognition sequence, and the downstream primer plus the XhoI recognition sequence (the underlined part is the restriction enzyme recognition sequence):
[0038] Upstream primer F-2: 5′-CGC GAGCTC ATGACACTTGAAAAATTTGTGGATGC-3' (SEQ ID NO. 5),
[0039] Downstream primer R-2: 5′-CCG CTCGAG TTATTTATGGGGATCAGTTATATC-3' (SEQ ID NO. 6),
[0040] Add each component according to the following PCR system to amplify the LOX gene:
[0041]
[0042] The PCR program is: 94°C for 3min; 30×(94°C for 40s; 53°C for 50s; 72°C for 90s); 72°C for 10min.
[0043] Purify the PCR product with Shanghai Sangon PCR Product Purification Kit, add SacI, XhoI double enzyme digestion, inactivation, ethanol precipitation, ddH 2 O was redissolved, ligated with an appropriate amount of vector pET-23a digested with the same restriction enzymes, and transformed into Escherichia coli DH5α. ...
Embodiment 3
[0045] Transform the expression plasmid pET-23a-fmb-L103 containing fmb-L103 into Escherichia coli expression host strain BL21(DE3)pLysS, culture at 37°C for 10-11 hours, pick small colonies, insert into 50ml LB liquid culture containing ampicillin Base, 70-90rpm, 30°C culture overnight, according to the volume ratio of 1:40, take the seed solution and add it to 100ml LB liquid medium containing ampicillin, shake at 180rpm at 35°C for 2-3 hours until the OD600 is about 0.6, add IPTG (final Concentration 100μg / ml) induced. After 1.5 hours, the cells were collected by centrifugation. The bacteria were crushed, and the supernatant was collected by centrifugation to obtain the crude enzyme solution of recombinant Bacillus vallismortis laccase (fmb-rL103).
[0046] The laccase activity was determined using the 2,2'-azino-bis(3-ethylbenzothiazole-6-sulfonic acid) (ABTS) method with slight modifications: the total volume of the reaction system was 3mL, including 2.45mL 0.2mol / L pH ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com